Distinct microbial communities associated with buried soils in the Siberian tundra
- PMID: 24335828
- PMCID: PMC3960545
- DOI: 10.1038/ismej.2013.219
Distinct microbial communities associated with buried soils in the Siberian tundra
Abstract
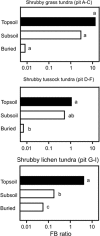
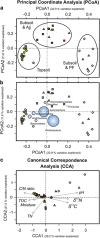
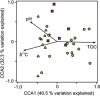
Cryoturbation, the burial of topsoil material into deeper soil horizons by repeated freeze-thaw events, is an important storage mechanism for soil organic matter (SOM) in permafrost-affected soils. Besides abiotic conditions, microbial community structure and the accessibility of SOM to the decomposer community are hypothesized to control SOM decomposition and thus have a crucial role in SOM accumulation in buried soils. We surveyed the microbial community structure in cryoturbated soils from nine soil profiles in the northeastern Siberian tundra using high-throughput sequencing and quantification of bacterial, archaeal and fungal marker genes. We found that bacterial abundances in buried topsoils were as high as in unburied topsoils. In contrast, fungal abundances decreased with depth and were significantly lower in buried than in unburied topsoils resulting in remarkably low fungal to bacterial ratios in buried topsoils. Fungal community profiling revealed an associated decrease in presumably ectomycorrhizal (ECM) fungi. The abiotic conditions (low to subzero temperatures, anoxia) and the reduced abundance of fungi likely provide a niche for bacterial, facultative anaerobic decomposers of SOM such as members of the Actinobacteria, which were found in significantly higher relative abundances in buried than in unburied topsoils. Our study expands the knowledge on the microbial community structure in soils of Northern latitude permafrost regions, and attributes the delayed decomposition of SOM in buried soils to specific microbial taxa, and particularly to a decrease in abundance and activity of ECM fungi, and to the extent to which bacterial decomposers are able to act as their functional substitutes.
Figures



References
-
- Abarenkov K, Henrik Nilsson R, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, et al. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 2010;186:281–285. - PubMed
-
- Aitchison J. The statistical analysis of compositional data. J R Stat Soc Ser B (Methodol) 1982;44:139–177.
-
- Allison SD. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett. 2005;8:626–635.
-
- Bailey VL, Smith JL, Bolton H., Jr Fungal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Bio Biochem. 2002;34:997–1007.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

