Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis
- PMID: 24336224
- PMCID: PMC4103798
- DOI: 10.1038/nsmb.2735
Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis
Abstract
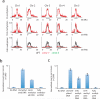
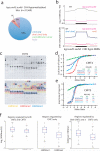
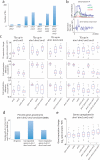
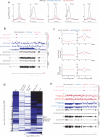
DNA methylation occurs in CG and non-CG sequence contexts. Non-CG methylation is abundant in plants and is mediated by CHROMOMETHYLASE (CMT) and DOMAINS REARRANGED METHYLTRANSFERASE (DRM) proteins; however, its roles remain poorly understood. Here we characterize the roles of non-CG methylation in Arabidopsis thaliana. We show that a poorly characterized methyltransferase, CMT2, is a functional methyltransferase in vitro and in vivo. CMT2 preferentially binds histone H3 Lys9 (H3K9) dimethylation and methylates non-CG cytosines that are regulated by H3K9 methylation. We revealed the contributions and redundancies between each non-CG methyltransferase in DNA methylation patterning and in regulating transcription. We also demonstrate extensive dependencies of small-RNA accumulation and H3K9 methylation patterning on non-CG methylation, suggesting self-reinforcing mechanisms between these epigenetic factors. The results suggest that non-CG methylation patterns are critical in shaping the landscapes of histone modification and small noncoding RNA.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

