Molecular signatures of antibody responses derived from a systems biology study of five human vaccines
- PMID: 24336226
- PMCID: PMC3946932
- DOI: 10.1038/ni.2789
Molecular signatures of antibody responses derived from a systems biology study of five human vaccines
Abstract
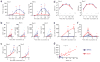
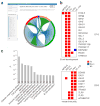
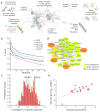
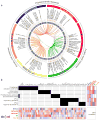
Many vaccines induce protective immunity via antibodies. Systems biology approaches have been used to determine signatures that can be used to predict vaccine-induced immunity in humans, but whether there is a 'universal signature' that can be used to predict antibody responses to any vaccine is unknown. Here we did systems analyses of immune responses to the polysaccharide and conjugate vaccines against meningococcus in healthy adults, in the broader context of published studies of vaccines against yellow fever virus and influenza virus. To achieve this, we did a large-scale network integration of publicly available human blood transcriptomes and systems-scale databases in specific biological contexts and deduced a set of transcription modules in blood. Those modules revealed distinct transcriptional signatures of antibody responses to different classes of vaccines, which provided key insights into primary viral, protein recall and anti-polysaccharide responses. Our results elucidate the early transcriptional programs that orchestrate vaccine immunity in humans and demonstrate the power of integrative network modeling.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Strength in numbers: comparing vaccine signatures the modular way.Nat Immunol. 2014 Feb;15(2):139-41. doi: 10.1038/ni.2807. Nat Immunol. 2014. PMID: 24448573 No abstract available.
References
-
- Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nature Reviews Immunology. 2009;9:741–747. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R37 AI048638/AI/NIAID NIH HHS/United States
- R38 AI140299/AI/NIAID NIH HHS/United States
- U19 AI090023/AI/NIAID NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- R37DK057665/DK/NIDDK NIH HHS/United States
- U19AI090023/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- U19 AI057266/AI/NIAID NIH HHS/United States
- R37 DK057665/DK/NIDDK NIH HHS/United States
- AI100663-02/AI/NIAID NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- U19 AI096187/AI/NIAID NIH HHS/United States
- R37AI48638/AI/NIAID NIH HHS/United States
- UL1 RR025008/RR/NCRR NIH HHS/United States
- U54 AI057157/AI/NIAID NIH HHS/United States
- R01 DK057665/DK/NIDDK NIH HHS/United States
- U19AI057266/AI/NIAID NIH HHS/United States
- P51 RR000165/RR/NCRR NIH HHS/United States
- U54AI057157/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

