Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin
- PMID: 24336483
- PMCID: PMC3863819
- DOI: 10.1038/srep03510
Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin
Erratum in
-
Corrigendum: Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin.Sci Rep. 2015 Jul 13;5:9628. doi: 10.1038/srep09628. Sci Rep. 2015. PMID: 26166249 Free PMC article. No abstract available.
Abstract
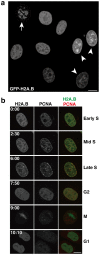
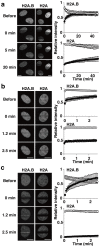
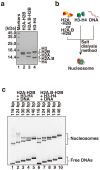
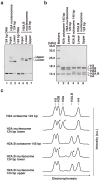
Human histone H2A.B (formerly H2A.Bbd), a non-allelic H2A variant, exchanges rapidly as compared to canonical H2A, and preferentially associates with actively transcribed genes. We found that H2A.B transiently accumulated at DNA replication and repair foci in living cells. To explore the biochemical function of H2A.B, we performed nucleosome reconstitution analyses using various lengths of DNA. Two types of H2A.B nucleosomes, octasome and hexasome, were formed with 116, 124, or 130 base pairs (bp) of DNA, and only the octasome was formed with 136 or 146 bp DNA. In contrast, only hexasome formation was observed by canonical H2A with 116 or 124 bp DNA. A small-angle X-ray scattering analysis revealed that the H2A.B octasome is more extended, due to the flexible detachment of the DNA regions at the entry/exit sites from the histone surface. These results suggested that H2A.B rapidly and transiently forms nucleosomes with short DNA segments during chromatin reorganization.
Figures
References
-
- Luger K., Mader A. W., Richmond R. K., Sargent D. F. & Richmond T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). - PubMed
-
- Davey C. A., Sargent D. F., Luger K., Maeder A. W. & Richmond T. J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113 (2002). - PubMed
-
- Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA Repair 4, 939–950 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

