Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce
- PMID: 24336492
- PMCID: PMC3904711
- DOI: 10.1093/jxb/ert398
Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce
Abstract
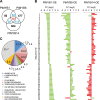
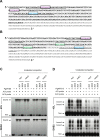
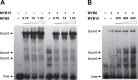
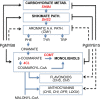
Redundancy and competition between R2R3-MYB activators and repressors on common target genes has been proposed as a fine-tuning mechanism for the regulation of plant secondary metabolism. This hypothesis was tested in white spruce [Picea glauca (Moench) Voss] by investigating the effects of R2R3-MYBs from different subgroups on common targets from distinct metabolic pathways. Comparative analysis of transcript profiling data in spruces overexpressing R2R3-MYBs from loblolly pine (Pinus taeda L.), PtMYB1, PtMYB8, and PtMYB14, defined a set of common genes that display opposite regulation effects. The relationship between the closest MYB homologues and 33 putative target genes was explored by quantitative PCR expression profiling in wild-type P. glauca plants during the diurnal cycle. Significant Spearman's correlation estimates were consistent with the proposed opposite effect of different R2R3-MYBs on several putative target genes in a time-related and tissue-preferential manner. Expression of sequences coding for 4CL, DHS2, COMT1, SHM4, and a lipase thio/esterase positively correlated with that of PgMYB1 and PgMYB8, but negatively with that of PgMYB14 and PgMYB15. Complementary electrophoretic mobility shift assay (EMSA) and transactivation assay provided experimental evidence that these different R2R3-MYBs are able to bind similar AC cis-elements in the promoter region of Pg4CL and PgDHS2 genes but have opposite effects on their expression. Competitive binding EMSA experiments showed that PgMYB8 competes more strongly than PgMYB15 for the AC-I MYB binding site in the Pg4CL promoter. Together, the results bring a new perspective to the action of R2R3-MYB proteins in the regulation of distinct but interconnecting metabolism pathways.
Keywords: Conifers; R2R3-MYB evolution; phenylpropanoid pathway; protein–DNA binding; transcriptional network..
Figures
References
-
- Bedon F, Levasseur C, Grima-Pettenati J, Séguin A, MacKay J. 2009. Sequence analysis and functional characterization of the promoter of the Picea glauca Cinnamyl Alcohol Dehydrogenase gene in transgenic white spruce plants. Plant Cell Reports 28, 787–800 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

