Genetic screens in human cells using the CRISPR-Cas9 system
- PMID: 24336569
- PMCID: PMC3972032
- DOI: 10.1126/science.1246981
Genetic screens in human cells using the CRISPR-Cas9 system
Abstract
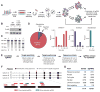
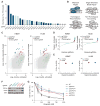
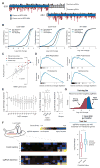
The bacterial clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system for genome editing has greatly expanded the toolbox for mammalian genetics, enabling the rapid generation of isogenic cell lines and mice with modified alleles. Here, we describe a pooled, loss-of-function genetic screening approach suitable for both positive and negative selection that uses a genome-scale lentiviral single-guide RNA (sgRNA) library. sgRNA expression cassettes were stably integrated into the genome, which enabled a complex mutant pool to be tracked by massively parallel sequencing. We used a library containing 73,000 sgRNAs to generate knockout collections and performed screens in two human cell lines. A screen for resistance to the nucleotide analog 6-thioguanine identified all expected members of the DNA mismatch repair pathway, whereas another for the DNA topoisomerase II (TOP2A) poison etoposide identified TOP2A, as expected, and also cyclin-dependent kinase 6, CDK6. A negative selection screen for essential genes identified numerous gene sets corresponding to fundamental processes. Last, we show that sgRNA efficiency is associated with specific sequence motifs, enabling the prediction of more effective sgRNAs. Collectively, these results establish Cas9/sgRNA screens as a powerful tool for systematic genetic analysis in mammalian cells.
Figures
References
-
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. - PubMed
-
- Carette JE, et al. Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens. Science. 2009;326:1231–1235. - PubMed
-
- Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. - PubMed
-
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

