Intergenic transcriptional interference is blocked by RNA polymerase III transcription factor TFIIIB in Saccharomyces cerevisiae
- PMID: 24336746
- PMCID: PMC3914616
- DOI: 10.1534/genetics.113.160093
Intergenic transcriptional interference is blocked by RNA polymerase III transcription factor TFIIIB in Saccharomyces cerevisiae
Abstract
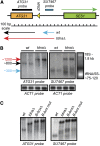
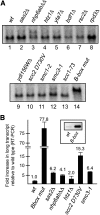
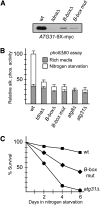
The major function of eukaryotic RNA polymerase III is to transcribe transfer RNA, 5S ribosomal RNA, and other small non-protein-coding RNA molecules. Assembly of the RNA polymerase III complex on chromosomal DNA requires the sequential binding of transcription factor complexes TFIIIC and TFIIIB. Recent evidence has suggested that in addition to producing RNA transcripts, chromatin-assembled RNA polymerase III complexes may mediate additional nuclear functions that include chromatin boundary, nucleosome phasing, and general genome organization activities. This study provides evidence of another such "extratranscriptional" activity of assembled RNA polymerase III complexes, which is the ability to block progression of intergenic RNA polymerase II transcription. We demonstrate that the RNA polymerase III complex bound to the tRNA gene upstream of the Saccharomyces cerevisiae ATG31 gene protects the ATG31 promoter against readthrough transcriptional interference from the upstream noncoding intergenic SUT467 transcription unit. This protection is predominately mediated by binding of the TFIIIB complex. When TFIIIB binding to this tRNA gene is weakened, an extended SUT467-ATG31 readthrough transcript is produced, resulting in compromised ATG31 translation. Since the ATG31 gene product is required for autophagy, strains expressing the readthrough transcript exhibit defective autophagy induction and reduced fitness under autophagy-inducing nitrogen starvation conditions. Given the recent discovery of widespread pervasive transcription in all forms of life, protection of neighboring genes from intergenic transcriptional interference may be a key extratranscriptional function of assembled RNA polymerase III complexes and possibly other DNA binding proteins.
Keywords: RNA polymerase III; TFIIIB; TFIIIC; extratranscriptional effects; noncoding transcription.
Figures


References
-
- Acker J., Conesa C., Lefebvre O., 2013. Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta 1829: 283–295. - PubMed
-
- Andrau J. C., Sentenac A., Werner M., 1999. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 288: 511–520. - PubMed
-
- Bardeleben C., Kassavetis G. A., Geiduschek E. P., 1994. Encounters of Saccharomyces cerevisiae RNA polymerase III with its transcription factors during RNA chain elongation. J. Mol. Biol. 235: 1193–1205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

