Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge
- PMID: 24336749
- PMCID: PMC3914621
- DOI: 10.1534/genetics.113.160531
Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge
Abstract
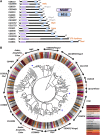
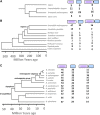
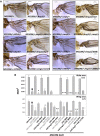
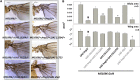
Gene duplication, expansion, and subsequent diversification are features of the evolutionary process. Duplicated genes can be lost, modified, or altered to generate novel functions over evolutionary timescales. These features make gene duplication a powerful engine of evolutionary change. In this study, we explore these features in the MADF-BESS family of transcriptional regulators. In Drosophila melanogaster, the family contains 16 similar members, each containing an N-terminal, DNA-binding MADF domain and a C-terminal, protein-interacting, BESS domain. Phylogenetic analysis shows that members of the MADF-BESS family are expanded in the Drosophila lineage. Three members, which we name hinge1, hinge2, and hinge3 are required for wing development, with a critical role in the wing hinge. hinge1 is a negative regulator of Winglesss expression and interacts with core wing-hinge patterning genes such as teashirt, homothorax, and jing. Double knockdowns along with heterologous rescue experiments are used to demonstrate that members of the MADF-BESS family retain function in the wing hinge, in spite of expansion and diversification for over 40 million years. The wing hinge connects the blade to the thorax and has critical roles in fluttering during flight. MADF-BESS family genes appear to retain redundant functions to shape and form elements of the wing hinge in a robust and fail-safe manner.
Keywords: BESS; MADF; Wnt/Wg; development; evolution.
Figures


Similar articles
-
JAK/STAT signaling is required for hinge growth and patterning in the Drosophila wing disc.Dev Biol. 2013 Oct 15;382(2):413-26. doi: 10.1016/j.ydbio.2013.08.016. Epub 2013 Aug 23. Dev Biol. 2013. PMID: 23978534 Free PMC article.
-
A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila.Development. 2000 Apr;127(7):1499-508. doi: 10.1242/dev.127.7.1499. Development. 2000. PMID: 10704395
-
The MADF-BESS Protein CP60 Is Recruited to Insulators via CP190 and Has Redundant Functions in Drosophila.Int J Mol Sci. 2023 Oct 9;24(19):15029. doi: 10.3390/ijms241915029. Int J Mol Sci. 2023. PMID: 37834476 Free PMC article.
-
Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing.Development. 2006 Jan;133(2):183-93. doi: 10.1242/dev.02214. Development. 2006. PMID: 16368928 Free PMC article. Review.
-
T-Box Genes in Drosophila Limb Development.Curr Top Dev Biol. 2017;122:313-354. doi: 10.1016/bs.ctdb.2016.08.003. Epub 2016 Sep 9. Curr Top Dev Biol. 2017. PMID: 28057269 Review.
Cited by
-
A pupal transcriptomic screen identifies Ral as a target of store-operated calcium entry in Drosophila neurons.Sci Rep. 2017 Feb 14;7:42586. doi: 10.1038/srep42586. Sci Rep. 2017. PMID: 28195208 Free PMC article.
-
Stonewall prevents expression of ectopic genes in the ovary and accumulates at insulator elements in D. melanogaster.PLoS Genet. 2022 Mar 24;18(3):e1010110. doi: 10.1371/journal.pgen.1010110. eCollection 2022 Mar. PLoS Genet. 2022. PMID: 35324887 Free PMC article.
-
New Drosophila promoter-associated architectural protein Mzfp1 interacts with CP190 and is required for housekeeping gene expression and insulator activity.Nucleic Acids Res. 2024 Jul 8;52(12):6886-6905. doi: 10.1093/nar/gkae393. Nucleic Acids Res. 2024. PMID: 38769058 Free PMC article.
-
PIF/harbinger transposon-derived protein promotes 7SL expression to enhance pathogen resistance.EMBO Rep. 2025 Mar;26(5):1196-1211. doi: 10.1038/s44319-025-00379-8. Epub 2025 Jan 30. EMBO Rep. 2025. PMID: 39885293 Free PMC article.
-
Evolution of the Drosophila melanogaster Chromatin Landscape and Its Associated Proteins.Genome Biol Evol. 2019 Mar 1;11(3):660-677. doi: 10.1093/gbe/evz019. Genome Biol Evol. 2019. PMID: 30689829 Free PMC article.
References
-
- Azpiazu N., Morata G., 2000. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development 127: 2685–2693. - PubMed
-
- Bhaskar V., Courey A. J., 2002. The MADF-BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene 299: 173–184. - PubMed
-
- Boyer L. A., Langer M. R., Crowley K. A., Tan S., Denu J. M., et al. , 2002. Essential role for the SANT domain chromatin remodeling enzymes. Mol. Cell 10: 935–942. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

