Feedback regulation of SIN by Etd1 and Rho1 in fission yeast
- PMID: 24336750
- PMCID: PMC3914619
- DOI: 10.1534/genetics.113.155218
Feedback regulation of SIN by Etd1 and Rho1 in fission yeast
Abstract
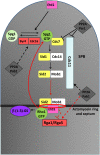
In fission yeast, the septation initiation network (SIN) is thought to promote cytokinesis by downstream activation of Rho1, a conserved GTPase that controls cell growth and division. Here we show that Etd1 and PP2A-Pab1, antagonistic regulators of SIN, are Rho1 regulators. Our genetic and biochemical studies indicate that a C-terminal region of Etd1 may activate Rho1 by directly binding it, whereas an N-terminal domain confers its ability to localize at the growing tips and the division site where Rho1 functions. In opposition to Etd1, our results indicate that PP2A-Pab1 inhibits Rho1. The SIN cascade is upstream-regulated by the Spg1 GTPase. In the absence of Etd1, activity of Spg1 drops down prematurely, thereby inactivating SIN. Interestingly, we find that ectopic activation of Rho1 restores Spg1 activity in Etd1-depleted cells. By using a cytokinesis block strategy, we show that Rho1 is essential to feedback-activate Spg1 during actomyosin ring constriction. Therefore, activation of Spg1 by Rho1, which in turn is regulated by Etd1, uncovers a novel feedback loop mechanism that ensures SIN activity while cytokinesis is progressing.
Keywords: Etd1; PP2A-Pab1; Rho1; SIN; Schizosaccharomyces pombe; Spg1; cytokinesis.
Figures

Similar articles
-
Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast.Genetics. 2010 Dec;186(4):1261-70. doi: 10.1534/genetics.110.121368. Epub 2010 Sep 27. Genetics. 2010. PMID: 20876564 Free PMC article.
-
Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1.J Cell Biol. 2009 Sep 7;186(5):739-53. doi: 10.1083/jcb.200902116. J Cell Biol. 2009. PMID: 19736319 Free PMC article.
-
Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis.Mol Biol Cell. 2008 Apr;19(4):1727-38. doi: 10.1091/mbc.e07-07-0718. Epub 2008 Feb 6. Mol Biol Cell. 2008. PMID: 18256290 Free PMC article.
-
The Multiple Functions of Rho GTPases in Fission Yeasts.Cells. 2021 Jun 7;10(6):1422. doi: 10.3390/cells10061422. Cells. 2021. PMID: 34200466 Free PMC article. Review.
-
Cell division: SIN, cytokinesis and ethanol dependency.Curr Biol. 2005 Aug 9;15(15):R605-7. doi: 10.1016/j.cub.2005.07.045. Curr Biol. 2005. PMID: 16085485 Review.
Cited by
-
Ser/Thr protein phosphatases in fungi: structure, regulation and function.Microb Cell. 2019 Apr 24;6(5):217-256. doi: 10.15698/mic2019.05.677. Microb Cell. 2019. PMID: 31114794 Free PMC article. Review.
-
The price of independence: cell separation in fission yeast.World J Microbiol Biotechnol. 2016 Apr;32(4):65. doi: 10.1007/s11274-016-2021-8. Epub 2016 Mar 1. World J Microbiol Biotechnol. 2016. PMID: 26931605 Review.
-
Cdc42 prevents precocious Rho1 activation during cytokinesis in a Pak1-dependent manner.J Cell Sci. 2023 Apr 15;136(8):jcs261160. doi: 10.1242/jcs.261160. Epub 2023 Apr 26. J Cell Sci. 2023. PMID: 37039135 Free PMC article.
-
Spatial control of secretory vesicle targeting by the Ync13-Rga7-Rng10 complex during cytokinesis.bioRxiv [Preprint]. 2025 Jul 9:2025.05.13.653810. doi: 10.1101/2025.05.13.653810. bioRxiv. 2025. PMID: 40463202 Free PMC article. Preprint.
-
Spatial control of translation repression and polarized growth by conserved NDR kinase Orb6 and RNA-binding protein Sts5.Elife. 2016 Jul 30;5:e14216. doi: 10.7554/eLife.14216. Elife. 2016. PMID: 27474797 Free PMC article.
References
-
- Arellano M., Duran A., Perez P., 1997. Localisation of the Schizosaccharomyces pombe rho1p GTPase and its involvement in the organisation of the actin cytoskeleton. J. Cell Sci. 110: 2547–2555. - PubMed
-
- Bardin A. J., Visintin R., Amon A., 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31. - PubMed
-
- Calonge T. M., Arellano M., Coll P. M., Perez P., 2003. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 47: 507–518. - PubMed
-
- Cerutti L., Simanis V., 1999. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J. Cell Sci. 112: 2313–2321. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

