Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit
- PMID: 24336829
- PMCID: PMC3922217
- DOI: 10.1093/cid/cit807
Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit
Abstract
Background: Strategies to prevent Staphylococcus aureus infection in hospitals focus on patient-to-patient transmission. We used whole-genome sequencing to investigate the role of colonized patients as the source of new S. aureus acquisitions, and the reliability of identifying patient-to-patient transmission using the conventional approach of spa typing and overlapping patient stay.
Methods: Over 14 months, all unselected patients admitted to an adult intensive care unit (ICU) were serially screened for S. aureus. All available isolates (n = 275) were spa typed and underwent whole-genome sequencing to investigate their relatedness at high resolution.
Results: Staphylococcus aureus was carried by 185 of 1109 patients sampled within 24 hours of ICU admission (16.7%); 59 (5.3%) patients carried methicillin-resistant S. aureus (MRSA). Forty-four S. aureus (22 MRSA) acquisitions while on ICU were detected. Isolates were available for genetic analysis from 37 acquisitions. Whole-genome sequencing indicated that 7 of these 37 (18.9%) were transmissions from other colonized patients. Conventional methods (spa typing combined with overlapping patient stay) falsely identified 3 patient-to-patient transmissions (all MRSA) and failed to detect 2 acquisitions and 4 transmissions (2 MRSA).
Conclusions: Only a minority of S. aureus acquisitions can be explained by patient-to-patient transmission. Whole-genome sequencing provides the resolution to disprove transmission events indicated by conventional methods and also to reveal otherwise unsuspected transmission events. Whole-genome sequencing should replace conventional methods for detection of nosocomial S. aureus transmission.
Keywords: Staphylococcus aureus transmission; adult; intensive care unit; spa typing; whole-genome sequencing.
Figures



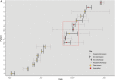
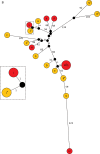
 ) or discordant spa types (
) or discordant spa types ( ).
).
Comment in
-
Applying a new technology to an old question: whole-genome sequencing and Staphylococcus aureus acquisition in an intensive care unit.Clin Infect Dis. 2014 Mar;58(5):619-21. doi: 10.1093/cid/cit812. Epub 2013 Dec 12. Clin Infect Dis. 2014. PMID: 24336830 No abstract available.
-
Data on whole-genome sequencing are insufficient to rule out patient-to-patient transmission as a significant source of Staphylococcus aureus acquisition in an intensive care unit.Clin Infect Dis. 2014 Sep 1;59(5):752. doi: 10.1093/cid/ciu368. Epub 2014 May 20. Clin Infect Dis. 2014. PMID: 24850799 No abstract available.
-
Reply to Mills and Linkin.Clin Infect Dis. 2014 Sep 1;59(5):752-3. doi: 10.1093/cid/ciu370. Epub 2014 May 20. Clin Infect Dis. 2014. PMID: 24850802 Free PMC article. No abstract available.
References
-
- Wertheim HF, Vos MC, Ott A, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet. 2004;364:703–5. - PubMed
-
- Wertheim HF, Melles DC, Vos MC, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–62. - PubMed
-
- Pearson A, Chronias A, Murray M. Voluntary and mandatory surveillance for methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) bacteraemia in England. J Antimicrob Chemother. 2009;64(suppl 1):i11–i7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

