The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes
- PMID: 24336862
- PMCID: PMC4072906
- DOI: 10.1093/jhered/est084
The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes
Abstract
Over 95% of all metazoan (animal) species comprise the "invertebrates," but very few genomes from these organisms have been sequenced. We have, therefore, formed a "Global Invertebrate Genomics Alliance" (GIGA). Our intent is to build a collaborative network of diverse scientists to tackle major challenges (e.g., species selection, sample collection and storage, sequence assembly, annotation, analytical tools) associated with genome/transcriptome sequencing across a large taxonomic spectrum. We aim to promote standards that will facilitate comparative approaches to invertebrate genomics and collaborations across the international scientific community. Candidate study taxa include species from Porifera, Ctenophora, Cnidaria, Placozoa, Mollusca, Arthropoda, Echinodermata, Annelida, Bryozoa, and Platyhelminthes, among others. GIGA will target 7000 noninsect/nonnematode species, with an emphasis on marine taxa because of the unrivaled phyletic diversity in the oceans. Priorities for selecting invertebrates for sequencing will include, but are not restricted to, their phylogenetic placement; relevance to organismal, ecological, and conservation research; and their importance to fisheries and human health. We highlight benefits of sequencing both whole genomes (DNA) and transcriptomes and also suggest policies for genomic-level data access and sharing based on transparency and inclusiveness. The GIGA Web site (http://giga.nova.edu) has been launched to facilitate this collaborative venture.
Keywords: GIGA; biodiversity; comparative genomics; consortium; evolution; invertebrates; metazoa.
Figures

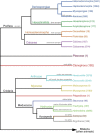
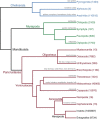
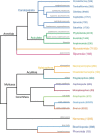

References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster . Science. 287:2185–2195 - PubMed
-
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. 1997. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 387:489–493 - PubMed
-
- Ahyong ST, Lowry JK, Alonso M, Bamber RN, Boxshall GA, Castro P, Gerken S, Karaman GS, Goy JW, Jones DS, et al. 2011. Subphylum Crustacea Brünnich, 1772. In: Zhang ZQ, editor. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Vol. 3148 Zootaxa; p. 165–191
-
- Appeltans W, Ahyong ST, Anderson G, Angel MV, Artois T, Bailly N, Bamber R, Barber A, Bartsch I, Berta A, et al. 2012. The magnitude of global marine species diversity. Curr Biol. 22:2189–2202 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

