Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation
- PMID: 24338021
- PMCID: PMC3908403
- DOI: 10.1074/jbc.M113.524926
Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation
Abstract
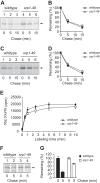
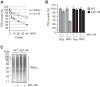
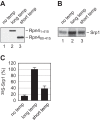
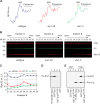
Cotranslational protein degradation plays an important role in protein quality control and proteostasis. Although ubiquitylation has been suggested to signal cotranslational degradation of nascent polypeptides, cotranslational ubiquitylation occurs at a low level, suggesting the existence of an alternative route for delivery of nascent polypeptides to the proteasome. Here we report that the nuclear import factor Srp1 (also known as importin α or karyopherin α) is required for ubiquitin-independent cotranslational degradation of the transcription factor Rpn4. We further demonstrate that cotranslational protein degradation is generally impaired in the srp1-49 mutant. Srp1 binds nascent polypeptides emerging from the ribosome. The association of proteasomes with polysomes is weakened in srp1-49. The interaction between Srp1 and the proteasome is mediated by Sts1, a multicopy suppressor of srp1-49. The srp1-49 and sts1-2 mutants are hypersensitive to stressors that promote protein misfolding, underscoring the physiological function of Srp1 and Sts1 in degradation of misfolded nascent polypeptides. This study unveils a previously unknown role for Srp1 and Sts1 in cotranslational protein degradation and suggests a novel model whereby Srp1 and Sts1 cooperate to couple proteasomes to ribosome-bound nascent polypeptides.
Keywords: Cotranslational Protein Degradation; Nuclear Import Factor Srp1; Proteasome; Protein Degradation; Protein Dynamics; Protein Processing; Protein Turnover; Ubiquitin-independent Degradation.
Figures


Similar articles
-
Yeast 26S proteasome nuclear import is coupled to nucleus-specific degradation of the karyopherin adaptor protein Sts1.Sci Rep. 2024 Jan 24;14(1):2048. doi: 10.1038/s41598-024-52352-5. Sci Rep. 2024. PMID: 38267508 Free PMC article.
-
The Sts1 nuclear import adapter uses a non-canonical bipartite nuclear localization signal and is directly degraded by the proteasome.J Cell Sci. 2020 Mar 19;133(6):jcs236158. doi: 10.1242/jcs.236158. J Cell Sci. 2020. PMID: 32041904 Free PMC article.
-
Sts1 plays a key role in targeting proteasomes to the nucleus.J Biol Chem. 2011 Jan 28;286(4):3104-18. doi: 10.1074/jbc.M110.135863. Epub 2010 Nov 12. J Biol Chem. 2011. PMID: 21075847 Free PMC article.
-
False start: cotranslational protein ubiquitination and cytosolic protein quality control.J Proteomics. 2014 Apr 4;100:92-101. doi: 10.1016/j.jprot.2013.08.005. Epub 2013 Aug 15. J Proteomics. 2014. PMID: 23954725 Review.
-
The ribosome-bound quality control complex: from aberrant peptide clearance to proteostasis maintenance.Curr Genet. 2017 Dec;63(6):997-1005. doi: 10.1007/s00294-017-0708-5. Epub 2017 May 20. Curr Genet. 2017. PMID: 28528489 Review.
Cited by
-
Nuclear transport of yeast proteasomes.Biomolecules. 2014 Oct 20;4(4):940-55. doi: 10.3390/biom4040940. Biomolecules. 2014. PMID: 25333764 Free PMC article. Review.
-
Translating DRiPs: MHC class I immunosurveillance of pathogens and tumors.J Leukoc Biol. 2014 Apr;95(4):551-62. doi: 10.1189/jlb.1113599. Epub 2014 Feb 14. J Leukoc Biol. 2014. PMID: 24532645 Free PMC article. Review.
-
Genotypic, proteomic, and phenotypic approaches to decipher the response to caspofungin and calcineurin inhibitors in clinical isolates of echinocandin-resistant Candida glabrata.J Antimicrob Chemother. 2022 Feb 23;77(3):585-597. doi: 10.1093/jac/dkab454. J Antimicrob Chemother. 2022. PMID: 34893830 Free PMC article.
-
Yeast 26S proteasome nuclear import is coupled to nucleus-specific degradation of the karyopherin adaptor protein Sts1.Sci Rep. 2024 Jan 24;14(1):2048. doi: 10.1038/s41598-024-52352-5. Sci Rep. 2024. PMID: 38267508 Free PMC article.
-
A nuclear ubiquitin-proteasome pathway targets the inner nuclear membrane protein Asi2 for degradation.J Cell Sci. 2014 Aug 15;127(Pt 16):3603-13. doi: 10.1242/jcs.153163. Epub 2014 Jun 13. J Cell Sci. 2014. PMID: 24928896 Free PMC article.
References
-
- Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 - PubMed
-
- Kramer G., Boehringer D., Ban N., Bukau B. (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 - PubMed
-
- Preissler S., Deuerling E. (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 - PubMed
-
- Tyedmers J., Mogk A., Bukau B. (2010) Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11, 777–788 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

