The regulation of the autophagic network and its implications for human disease
- PMID: 24339733
- PMCID: PMC3858585
- DOI: 10.7150/ijbs.6666
The regulation of the autophagic network and its implications for human disease
Abstract
Autophagy has attracted a lot of attention in recent years. More and more proteins and signaling pathways have been discovered that somehow feed into the autophagy regulatory pathways. Regulation of autophagy is complex and condition-specific, and in several diseases, autophagic fluxes are changed. Here, we review the most well-established concepts in this field as well as the reported signaling pathways or components which steer the autophagy machinery. Furthermore, we will highlight how autophagic fluxes are changed in various diseases either as cause for or as response to deal with an altered cellular homeostasis and how modulation of autophagy might be used as potential therapy for such diseases.
Keywords: ATGs; Autophagy; FOXO1; HSF1; Heat Shock Proteins (HSP); Human diseases.; UPR; mTOR.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
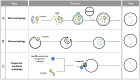
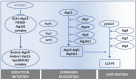
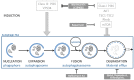
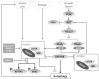
Similar articles
-
Heat shock factor 1 confers resistance to Hsp90 inhibitors through p62/SQSTM1 expression and promotion of autophagic flux.Biochem Pharmacol. 2014 Feb 1;87(3):445-55. doi: 10.1016/j.bcp.2013.11.014. Epub 2013 Nov 28. Biochem Pharmacol. 2014. PMID: 24291777 Free PMC article.
-
Regulation of autophagy by stress-responsive transcription factors.Semin Cancer Biol. 2013 Oct;23(5):310-22. doi: 10.1016/j.semcancer.2013.05.008. Epub 2013 May 30. Semin Cancer Biol. 2013. PMID: 23726895 Review.
-
Targeting autophagic pathways for cancer drug discovery.Chin J Cancer. 2013 Mar;32(3):113-20. doi: 10.5732/cjc.012.10010. Epub 2012 Jul 26. Chin J Cancer. 2013. PMID: 22835386 Free PMC article. Review.
-
Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: from pathophysiology to treatment.J Hematol Oncol. 2017 Mar 9;10(1):67. doi: 10.1186/s13045-017-0436-9. J Hematol Oncol. 2017. PMID: 28279189 Free PMC article. Review.
-
Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways.Ann N Y Acad Sci. 2020 May;1467(1):3-20. doi: 10.1111/nyas.14305. Epub 2020 Jan 27. Ann N Y Acad Sci. 2020. PMID: 31985829 Review.
Cited by
-
Friend or Foe: Paradoxical Roles of Autophagy in Gliomagenesis.Cells. 2021 Jun 6;10(6):1411. doi: 10.3390/cells10061411. Cells. 2021. PMID: 34204169 Free PMC article. Review.
-
WX20120108, a novel IAP antagonist, induces tumor cell autophagy via activating ROS-FOXO pathway.Acta Pharmacol Sin. 2019 Nov;40(11):1466-1479. doi: 10.1038/s41401-019-0253-5. Epub 2019 Jul 17. Acta Pharmacol Sin. 2019. PMID: 31316176 Free PMC article.
-
Regulation of alveolar macrophage death in pulmonary fibrosis: a review.Apoptosis. 2023 Dec;28(11-12):1505-1519. doi: 10.1007/s10495-023-01888-4. Epub 2023 Sep 14. Apoptosis. 2023. PMID: 37707713 Free PMC article. Review.
-
Imbalanced TGFβ signalling and autophagy drive erythroid priming of hematopoietic stem cells in β-thalassemia.Nat Commun. 2025 Jul 1;16(1):5639. doi: 10.1038/s41467-025-60676-7. Nat Commun. 2025. PMID: 40592819 Free PMC article.
-
COPD-Related Modification to the Airway Epithelium Permits Intracellular Residence of Nontypeable Haemophilus influenzae and May Be Potentiated by Macrolide Arrest of Autophagy.Int J Chron Obstruct Pulmon Dis. 2020 Jun 4;15:1253-1260. doi: 10.2147/COPD.S245819. eCollection 2020. Int J Chron Obstruct Pulmon Dis. 2020. PMID: 32581530 Free PMC article.
References
-
- Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12(7):665–75. - PubMed
-
- Nakatogawa H, Ohbayashi S, Sakoh-Nakatogawa M, Kakuta S, Suzuki SW, Kirisako H. et al. The Autophagy-related Protein Kinase Atg1 Interacts with the Ubiquitin-like Protein Atg8 via the Atg8 Family Interacting Motif to Facilitate Autophagosome Formation. J Biol Chem. 2012;287(34):28503–7. - PMC - PubMed
-
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–67. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

