Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation
- PMID: 24339747
- PMCID: PMC3858243
- DOI: 10.1371/journal.pbio.1001731
Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation
Abstract
Regulation of translation initiation is well appropriate to adapt cell growth in response to stress and environmental changes. Many bacterial mRNAs adopt structures in their 5' untranslated regions that modulate the accessibility of the 30S ribosomal subunit. Structured mRNAs interact with the 30S in a two-step process where the docking of a folded mRNA precedes an accommodation step. Here, we used a combination of experimental approaches in vitro (kinetic of mRNA unfolding and binding experiments to analyze mRNA-protein or mRNA-ribosome complexes, toeprinting assays to follow the formation of ribosomal initiation complexes) and in vivo (genetic) to monitor the action of ribosomal protein S1 on the initiation of structured and regulated mRNAs. We demonstrate that r-protein S1 endows the 30S with an RNA chaperone activity that is essential for the docking and the unfolding of structured mRNAs, and for the correct positioning of the initiation codon inside the decoding channel. The first three OB-fold domains of S1 retain all its activities (mRNA and 30S binding, RNA melting activity) on the 30S subunit. S1 is not required for all mRNAs and acts differently on mRNAs according to the signals present at their 5' ends. This work shows that S1 confers to the ribosome dynamic properties to initiate translation of a large set of mRNAs with diverse structural features.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
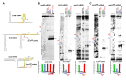


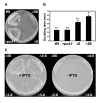
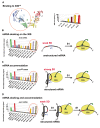
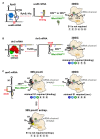
References
-
- Milón P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV (2012) Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol 19: 609–615. - PubMed
-
- Studer SM, Joseph S (2006) Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell 22: 105–115. - PubMed
-
- Yusupova G, Yusupov MM, Cate JH, Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106: 233–241. - PubMed
-
- Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, et al. (2005) Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science 308: 120–123. - PubMed
-
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M (2006) Structural basis for messenger RNA movement on the ribosome. Nature 444: 391–394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

