Circadian period integrates network information through activation of the BMP signaling pathway
- PMID: 24339749
- PMCID: PMC3858370
- DOI: 10.1371/journal.pbio.1001733
Circadian period integrates network information through activation of the BMP signaling pathway
Abstract
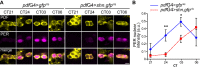
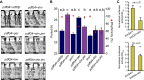
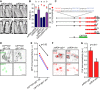
Living organisms use biological clocks to maintain their internal temporal order and anticipate daily environmental changes. In Drosophila, circadian regulation of locomotor behavior is controlled by ∼150 neurons; among them, neurons expressing the PIGMENT DISPERSING FACTOR (PDF) set the period of locomotor behavior under free-running conditions. To date, it remains unclear how individual circadian clusters integrate their activity to assemble a distinctive behavioral output. Here we show that the BONE MORPHOGENETIC PROTEIN (BMP) signaling pathway plays a crucial role in setting the circadian period in PDF neurons in the adult brain. Acute deregulation of BMP signaling causes period lengthening through regulation of dClock transcription, providing evidence for a novel function of this pathway in the adult brain. We propose that coherence in the circadian network arises from integration in PDF neurons of both the pace of the cell-autonomous molecular clock and information derived from circadian-relevant neurons through release of BMP ligands.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Sheeba V (2008) The Drosophila melanogaster circadian pacemaker circuit. J Genet 87: 485–493. - PubMed
-
- Helfrich-Forster C (1998) Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol A 182: 435–453. - PubMed
-
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

