Signatures of pleiotropy, economy and convergent evolution in a domain-resolved map of human-virus protein-protein interaction networks
- PMID: 24339775
- PMCID: PMC3855575
- DOI: 10.1371/journal.ppat.1003778
Signatures of pleiotropy, economy and convergent evolution in a domain-resolved map of human-virus protein-protein interaction networks
Abstract
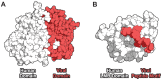
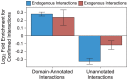
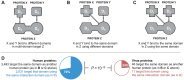
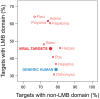
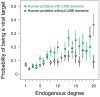
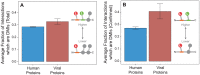
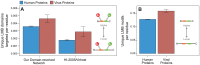
A central challenge in host-pathogen systems biology is the elucidation of general, systems-level principles that distinguish host-pathogen interactions from within-host interactions. Current analyses of host-pathogen and within-host protein-protein interaction networks are largely limited by their resolution, treating proteins as nodes and interactions as edges. Here, we construct a domain-resolved map of human-virus and within-human protein-protein interaction networks by annotating protein interactions with high-coverage, high-accuracy, domain-centric interaction mechanisms: (1) domain-domain interactions, in which a domain in one protein binds to a domain in a second protein, and (2) domain-motif interactions, in which a domain in one protein binds to a short, linear peptide motif in a second protein. Analysis of these domain-resolved networks reveals, for the first time, significant mechanistic differences between virus-human and within-human interactions at the resolution of single domains. While human proteins tend to compete with each other for domain binding sites by means of sequence similarity, viral proteins tend to compete with human proteins for domain binding sites in the absence of sequence similarity. Independent of their previously established preference for targeting human protein hubs, viral proteins also preferentially target human proteins containing linear motif-binding domains. Compared to human proteins, viral proteins participate in more domain-motif interactions, target more unique linear motif-binding domains per residue, and contain more unique linear motifs per residue. Together, these results suggest that viruses surmount genome size constraints by convergently evolving multiple short linear motifs in order to effectively mimic, hijack, and manipulate complex host processes for their survival. Our domain-resolved analyses reveal unique signatures of pleiotropy, economy, and convergent evolution in viral-host interactions that are otherwise hidden in the traditional binary network, highlighting the power and necessity of high-resolution approaches in host-pathogen systems biology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Alcami A (2003) Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol 3: 36–50. - PubMed
-
- Benedict CA, Norris PS, Ware CF (2002) To kill or be killed: viral evasion of apoptosis. Nat Immunol 3: 1013–1018. - PubMed
-
- Finlay BB, McFadden G (2006) Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124: 767–782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

