Basolateral Mg2+ extrusion via CNNM4 mediates transcellular Mg2+ transport across epithelia: a mouse model
- PMID: 24339795
- PMCID: PMC3854942
- DOI: 10.1371/journal.pgen.1003983
Basolateral Mg2+ extrusion via CNNM4 mediates transcellular Mg2+ transport across epithelia: a mouse model
Abstract
Transcellular Mg(2+) transport across epithelia, involving both apical entry and basolateral extrusion, is essential for magnesium homeostasis, but molecules involved in basolateral extrusion have not yet been identified. Here, we show that CNNM4 is the basolaterally located Mg(2+) extrusion molecule. CNNM4 is strongly expressed in intestinal epithelia and localizes to their basolateral membrane. CNNM4-knockout mice showed hypomagnesemia due to the intestinal malabsorption of magnesium, suggesting its role in Mg(2+) extrusion to the inner parts of body. Imaging analyses revealed that CNNM4 can extrude Mg(2+) by exchanging intracellular Mg(2+) with extracellular Na(+). Furthermore, CNNM4 mutations cause Jalili syndrome, characterized by recessive amelogenesis imperfecta with cone-rod dystrophy. CNNM4-knockout mice showed defective amelogenesis, and CNNM4 again localizes to the basolateral membrane of ameloblasts, the enamel-forming epithelial cells. Missense point mutations associated with the disease abolish the Mg(2+) extrusion activity. These results demonstrate the crucial importance of Mg(2+) extrusion by CNNM4 in organismal and topical regulation of magnesium.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
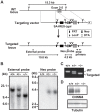
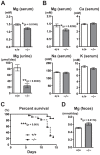

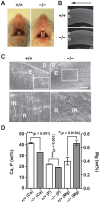
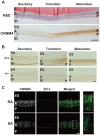
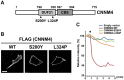
References
-
- Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, et al. (1999) Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103–106. - PubMed
-
- Hou J, Paul DL, Goodenough DA (2005) Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci 118: 5109–5118. - PubMed
-
- Schlingmann KP, Weber S, Peters M, Niemann Nejsum L, Vitzthum H, et al. (2002) Hypomagnesemia with secondary hypocalcemia is caused by mutations in TRPM6, a new member of the TRPM gene family. Nat Genet 31: 166–170. - PubMed
-
- Walder RY, Landau D, Meyer P, Shalev H, Tsolia M, et al. (2002) Mutation of TRPM6 causes familial hypomagnesemia with secondary hypocalcemia. Nat Genet 31: 171–174. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

