The proteasome inhibitor bortezomib induces an inhibitory chromatin environment at a distal enhancer of the estrogen receptor-α gene
- PMID: 24339902
- PMCID: PMC3855213
- DOI: 10.1371/journal.pone.0081110
The proteasome inhibitor bortezomib induces an inhibitory chromatin environment at a distal enhancer of the estrogen receptor-α gene
Abstract
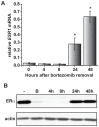
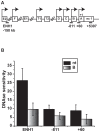
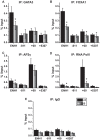
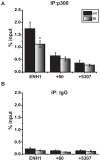
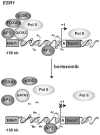
Expression of the estrogen receptor-α (ERα) gene, ESR1, is a clinical biomarker used to predict therapeutic outcome of breast cancer. Hence, there is significant interest in understanding the mechanisms regulating ESR1 gene expression. Proteasome activity is increased in cancer and we previously showed that proteasome inhibition leads to loss of ESR1 gene expression in breast cancer cells. Expression of ESR1 mRNA in breast cancer cells is controlled predominantly through a proximal promoter within ∼400 base pair (bp) of the transcription start site (TSS). Here, we show that loss of ESR1 gene expression induced by the proteasome inhibitor bortezomib is associated with inactivation of a distal enhancer located 150 kilobases (kb) from the TSS. Chromatin immunoprecipitation assays reveal several bortezomib-induced changes at the distal site including decreased occupancy of three critical transcription factors, GATA3, FOXA1, and AP2γ. Bortezomib treatment also resulted in decreased histone H3 and H4 acetylation and decreased occupancy of histone acetyltransferase, p300. These data suggest a mechanism to explain proteasome inhibitor-induced loss of ESR1 mRNA expression that highlights the importance of the chromatin environment at the -150 kb distal enhancer in regulation of basal expression of ESR1 in breast cancer cells.
Conflict of interest statement
Figures

Similar articles
-
Proteasome inhibition represses ERalpha gene expression in ER+ cells: a new link between proteasome activity and estrogen signaling in breast cancer.Oncogene. 2010 Mar 11;29(10):1509-18. doi: 10.1038/onc.2009.434. Epub 2009 Nov 30. Oncogene. 2010. PMID: 19946334 Free PMC article.
-
The proteasome inhibitor Bortezomib (Velcade) as potential inhibitor of estrogen receptor-positive breast cancer.Int J Cancer. 2015 Aug 1;137(3):686-97. doi: 10.1002/ijc.29404. Epub 2015 Jan 8. Int J Cancer. 2015. PMID: 25530422
-
Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B.Cancer Res. 2011 Sep 1;71(17):5739-53. doi: 10.1158/0008-5472.CAN-11-1896. Epub 2011 Aug 23. Cancer Res. 2011. PMID: 21862633
-
Positive Regulation of Estrogen Receptor Alpha in Breast Tumorigenesis.Cells. 2021 Oct 31;10(11):2966. doi: 10.3390/cells10112966. Cells. 2021. PMID: 34831189 Free PMC article. Review.
-
Proteasome inhibition in the treatment of cancer.Cell Cycle. 2005 Feb;4(2):290-6. Epub 2005 Feb 3. Cell Cycle. 2005. PMID: 15655370 Review.
Cited by
-
Prolonged unfolded protein reaction is involved in the induction of chronic myeloid leukemia cell death upon oprozomib treatment.Cancer Sci. 2021 Jan;112(1):133-143. doi: 10.1111/cas.14696. Epub 2020 Nov 12. Cancer Sci. 2021. PMID: 33067904 Free PMC article.
-
Bortezomib-Induced Epigenetic Alterations in Nerve Cells: Focus on the Mechanisms Contributing to the Peripheral Neuropathy Development.Int J Mol Sci. 2022 Feb 23;23(5):2431. doi: 10.3390/ijms23052431. Int J Mol Sci. 2022. PMID: 35269574 Free PMC article.
-
Intrinsic and Extrinsic Factors Governing the Transcriptional Regulation of ESR1.Horm Cancer. 2020 Aug;11(3-4):129-147. doi: 10.1007/s12672-020-00388-0. Epub 2020 Jun 26. Horm Cancer. 2020. PMID: 32592004 Free PMC article. Review.
-
Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation.Biomolecules. 2020 Mar 26;10(4):500. doi: 10.3390/biom10040500. Biomolecules. 2020. PMID: 32224970 Free PMC article. Review.
-
Estrogen Receptor Bio-Activities Determine Clinical Endocrine Treatment Options in Estrogen Receptor-Positive Breast Cancer.Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221090351. doi: 10.1177/15330338221090351. Technol Cancer Res Treat. 2022. PMID: 35450488 Free PMC article. Review.
References
-
- Cobleigh MA, Tabesh B, Bitterman P, Baker J, Cronin M, et al. (2005) Tumor Gene Expression and Prognosis in Breast Cancer Patients with 10 or More Positive Lymph Nodes. Clinical Cancer Research 11: 8623–8631. - PubMed
-
- Kos M, Reid G, Denger S, Gannon F (2001) Minireview: genomic organization of the human ERalpha gene promoter region. Mol Endocrinol 15: 2057–2063. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

