To control false positives in gene-gene interaction analysis: two novel conditional entropy-based approaches
- PMID: 24339984
- PMCID: PMC3858311
- DOI: 10.1371/journal.pone.0081984
To control false positives in gene-gene interaction analysis: two novel conditional entropy-based approaches
Abstract
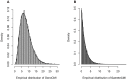
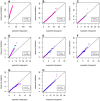
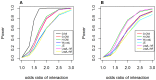
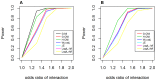
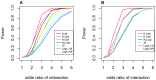
Genome-wide analysis of gene-gene interactions has been recognized as a powerful avenue to identify the missing genetic components that can not be detected by using current single-point association analysis. Recently, several model-free methods (e.g. the commonly used information based metrics and several logistic regression-based metrics) were developed for detecting non-linear dependence between genetic loci, but they are potentially at the risk of inflated false positive error, in particular when the main effects at one or both loci are salient. In this study, we proposed two conditional entropy-based metrics to challenge this limitation. Extensive simulations demonstrated that the two proposed metrics, provided the disease is rare, could maintain consistently correct false positive rate. In the scenarios for a common disease, our proposed metrics achieved better or comparable control of false positive error, compared to four previously proposed model-free metrics. In terms of power, our methods outperformed several competing metrics in a range of common disease models. Furthermore, in real data analyses, both metrics succeeded in detecting interactions and were competitive with the originally reported results or the logistic regression approaches. In conclusion, the proposed conditional entropy-based metrics are promising as alternatives to current model-based approaches for detecting genuine epistatic effects.
Conflict of interest statement
Figures







References
-
- Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 95–108. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

