A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates
- PMID: 24341331
- PMCID: PMC3878649
- DOI: 10.1186/1754-6834-6-182
A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates
Abstract
Background: Select cellulolytic bacteria produce multi-enzymatic cellulosome complexes that bind to the plant cell wall and catalyze its efficient degradation. The multi-modular interconnecting cellulosomal subunits comprise dockerin-containing enzymes that bind cohesively to cohesin-containing scaffoldins. The organization of the modules into functional polypeptides is achieved by intermodular linkers of different lengths and composition, which provide flexibility to the complex and determine its overall architecture.
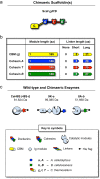
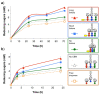
Results: Using a synthetic biology approach, we systematically investigated the spatial organization of the scaffoldin subunit and its effect on cellulose hydrolysis by designing a combinatorial library of recombinant trivalent designer scaffoldins, which contain a carbohydrate-binding module (CBM) and 3 divergent cohesin modules. The positions of the individual modules were shuffled into 24 different arrangements of chimaeric scaffoldins. This basic set was further extended into three sub-sets for each arrangement with intermodular linkers ranging from zero (no linkers), 5 (short linkers) and native linkers of 27-35 amino acids (long linkers). Of the 72 possible scaffoldins, 56 were successfully cloned and 45 of them expressed, representing 14 full sets of chimaeric scaffoldins. The resultant 42-component scaffoldin library was used to assemble designer cellulosomes, comprising three model C. thermocellum cellulases. Activities were examined using Avicel as a pure microcrystalline cellulose substrate and pretreated cellulose-enriched wheat straw as a model substrate derived from a native source. All scaffoldin combinations yielded active trivalent designer cellulosome assemblies on both substrates that exceeded the levels of the free enzyme systems. A preferred modular arrangement for the trivalent designer scaffoldin was not observed for the three enzymes used in this study, indicating that they could be integrated at any position in the designer cellulosome without significant effect on cellulose-degrading activity. Designer cellulosomes assembled with the long-linker scaffoldins achieved higher levels of activity, compared to those assembled with short-and no-linker scaffoldins.
Conclusions: The results demonstrate the robustness of the cellulosome system. Long intermodular scaffoldin linkers are preferable, thus leading to enhanced degradation of cellulosic substrates, presumably due to the increased flexibility and spatial positioning of the attached enzymes in the complex. These findings provide a general basis for improved designer cellulosome systems as a platform for bioethanol production.
Figures







Similar articles
-
Adaptor Scaffoldins: An Original Strategy for Extended Designer Cellulosomes, Inspired from Nature.mBio. 2016 Apr 5;7(2):e00083. doi: 10.1128/mBio.00083-16. mBio. 2016. PMID: 27048796 Free PMC article.
-
Deconstruction of lignocellulose into soluble sugars by native and designer cellulosomes.mBio. 2012 Dec 11;3(6):e00508-12. doi: 10.1128/mBio.00508-12. mBio. 2012. PMID: 23232718 Free PMC article.
-
Significance of relative position of cellulases in designer cellulosomes for optimized cellulolysis.PLoS One. 2015 May 29;10(5):e0127326. doi: 10.1371/journal.pone.0127326. eCollection 2015. PLoS One. 2015. PMID: 26024227 Free PMC article.
-
Cellulosomes: Highly Efficient Cellulolytic Complexes.Subcell Biochem. 2021;96:323-354. doi: 10.1007/978-3-030-58971-4_9. Subcell Biochem. 2021. PMID: 33252735 Review.
-
Cellulosomes-structure and ultrastructure.J Struct Biol. 1998 Dec 15;124(2-3):221-34. doi: 10.1006/jsbi.1998.4065. J Struct Biol. 1998. PMID: 10049808 Review.
Cited by
-
Pan-Cellulosomics of Mesophilic Clostridia: Variations on a Theme.Microorganisms. 2017 Nov 18;5(4):74. doi: 10.3390/microorganisms5040074. Microorganisms. 2017. PMID: 29156585 Free PMC article.
-
Enhancement of cellulosome-mediated deconstruction of cellulose by improving enzyme thermostability.Biotechnol Biofuels. 2016 Aug 4;9:164. doi: 10.1186/s13068-016-0577-z. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27493686 Free PMC article.
-
Toward combined delignification and saccharification of wheat straw by a laccase-containing designer cellulosome.Proc Natl Acad Sci U S A. 2016 Sep 27;113(39):10854-9. doi: 10.1073/pnas.1608012113. Epub 2016 Sep 12. Proc Natl Acad Sci U S A. 2016. PMID: 27621442 Free PMC article.
-
Minimalistic Cellulosome of the Butanologenic Bacterium Clostridium saccharoperbutylacetonicum.mBio. 2020 Mar 31;11(2):e00443-20. doi: 10.1128/mBio.00443-20. mBio. 2020. PMID: 32234813 Free PMC article.
-
Analysis of Engineered Tobacco Mosaic Virus and Potato Virus X Nanoparticles as Carriers for Biocatalysts.Front Plant Sci. 2021 Aug 6;12:710869. doi: 10.3389/fpls.2021.710869. eCollection 2021. Front Plant Sci. 2021. PMID: 34421958 Free PMC article.
References
-
- Lamed R, Setter E, Kenig R, Bayer EA. The cellulosome - a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol Bioeng Symp. 1983;6:163–181.
LinkOut - more resources
Full Text Sources
Other Literature Sources

