The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families
- PMID: 24341524
- PMCID: PMC6638672
- DOI: 10.1111/mpp.12099
The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families
Abstract
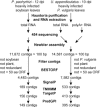
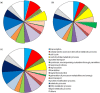
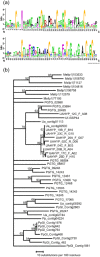
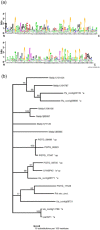
Haustoria of biotrophic rust fungi are responsible for the uptake of nutrients from their hosts and for the production of secreted proteins, known as effectors, which modulate the host immune system. The identification of the transcriptome of haustoria and an understanding of the functions of expressed genes therefore hold essential keys for the elucidation of fungus-plant interactions and the development of novel fungal control strategies. Here, we purified haustoria from infected leaves and used 454 sequencing to examine the haustorial transcriptomes of Phakopsora pachyrhizi and Uromyces appendiculatus, the causal agents of soybean rust and common bean rust, respectively. These pathogens cause extensive yield losses in their respective legume crop hosts. A series of analyses were used to annotate expressed sequences, including transposable elements and viruses, to predict secreted proteins from the assembled sequences and to identify families of candidate effectors. This work provides a foundation for the comparative analysis of haustorial gene expression with further insights into physiology and effector evolution.
© 2013 BSPP AND JOHN WILEY & SONS LTD.
Figures


References
-
- Apweiler, R. , Bairoch, A. , Wu, C.H. , Barker, W.C. , Boeckmann, B. , Ferro, S. , Gasteiger, E. , Huang, H. , Lopez, R. , Magrane, M. , Martin, M.J. , Natale, D.A. , O'Donovan, C. , Redaschi, N. and Yeh, L.S. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119. - PMC - PubMed
-
- Araya, C.M. , Alleyne, A.T. , Steadman, J.R. , Eskridgne, K.M. and Coyne, D.P. (2004) Phenotypic and genotypic characterization of Uromyces appendiculatus from Phaseolus vulgaris in the Americas. Plant Dis. 88, 830–836. - PubMed
-
- Bendtsen, J.D. , Nielsen, H. , von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. - PubMed
-
- Bonferroni, C.E. (1935) Ill calcolo delle assicurazioni su gruppi di teste. Studi in Onore del Professore Salvatore Ortu Carboni 13–60. Rome, Italy.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

