Ancient signals: comparative genomics of green plant CDPKs
- PMID: 24342084
- PMCID: PMC3932502
- DOI: 10.1016/j.tplants.2013.10.009
Ancient signals: comparative genomics of green plant CDPKs
Abstract
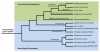
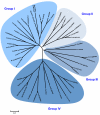
Calcium-dependent protein kinases (CDPKs) are multifunctional proteins that combine calcium-binding and signaling capabilities within a single gene product. This unique versatility enables multiple plant biological processes to be controlled, including developmental programs and stress responses. The genome of flowering plants typically encodes around 30 CDPK homologs that cluster in four conserved clades. In this review, we take advantage of the recent availability of genome sequences from green algae and early land plants to examine how well the previously described CDPK family from angiosperms compares to the broader evolutionary states associated with early diverging green plant lineages. Our analysis suggests that the current architecture of the CDPK family was shaped during the colonization of the land by plants, whereas CDPKs from ancestor green algae have continued to evolve independently.
Keywords: CDPKs; calcium signaling; evolution; green plant lineages.
Crown Copyright © 2013. Published by Elsevier Ltd. All rights reserved.
Figures


References
-
- Wheeler GL, Brownlee C. Ca2+ signalling in plants and green algae-changing channels. Trends Plant Sci. 2008;13:506–514. - PubMed
-
- Batistič O, Kudla J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta. 2012;1820:1283–1293. - PubMed
-
- Hashimoto K, Kudla J. Calcium decoding mechanisms in plants. Biochimie. 2011;93:2054–2059. - PubMed
-
- McCormack E, et al. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005;10:383–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

