Genomic analyses of DNA transformation and penicillin resistance in Streptococcus pneumoniae clinical isolates
- PMID: 24342643
- PMCID: PMC3957846
- DOI: 10.1128/AAC.01311-13
Genomic analyses of DNA transformation and penicillin resistance in Streptococcus pneumoniae clinical isolates
Abstract
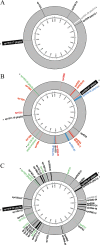
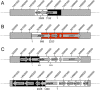
Alterations in penicillin-binding proteins, the target enzymes for β-lactam antibiotics, are recognized as primary penicillin resistance mechanisms in Streptococcus pneumoniae. Few studies have analyzed penicillin resistance at the genome scale, however, and we report the sequencing of S. pneumoniae R6 transformants generated while reconstructing the penicillin resistance phenotypes from three penicillin-resistant clinical isolates by serial genome transformation. The genome sequences of the three last-level transformants T2-18209, T5-1983, and T3-55938 revealed that 16.2 kb, 82.7 kb, and 137.2 kb of their genomes had been replaced with 5, 20, and 37 recombinant sequence segments derived from their respective parental clinical isolates, documenting the extent of DNA transformation between strains. A role in penicillin resistance was confirmed for some of the mutations identified in the transformants. Several multiple recombination events were also found to have happened at single loci coding for penicillin-binding proteins (PBPs) that increase resistance. Sequencing of the transformants with MICs for penicillin similar to those of the parent clinical strains confirmed the importance of mosaic PBP2x, -2b, and -1a as a driving force in penicillin resistance. A role in resistance for mosaic PBP2a was also observed for two of the resistant clinical isolates.
Figures

References
-
- Chesnel L, Carapito R, Croize J, Dideberg O, Vernet T, Zapun A. 2005. Identical penicillin-binding domains in penicillin-binding proteins of Streptococcus pneumoniae clinical isolates with different levels of beta-lactam resistance. Antimicrob. Agents Chemother. 49:2895–2902. 10.1128/AAC.49.7.2895-2902.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

