Dynamic regulation of transcriptional states by chromatin and transcription factors
- PMID: 24342920
- PMCID: PMC6322398
- DOI: 10.1038/nrg3623
Dynamic regulation of transcriptional states by chromatin and transcription factors
Abstract
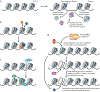
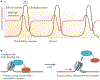
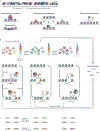
The interaction of regulatory proteins with the complex nucleoprotein structures that are found in mammalian cells involves chromatin reorganization at multiple levels. Mechanisms that support these transitions are complex on many timescales, which range from milliseconds to minutes or hours. In this Review, we discuss emerging concepts regarding the function of regulatory elements in living cells. We also explore the involvement of these dynamic and stochastic processes in the evolution of fluctuating transcriptional activity states that are now commonly reported in eukaryotic systems.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures

References
-
- Spitz F & Furlong EE Transcription factors: from enhancer binding to developmental control. Nature Rev. Genet 13, 613–626 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

