Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity
- PMID: 24344267
- PMCID: PMC3890789
- DOI: 10.1073/pnas.1320777110
Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity
Abstract
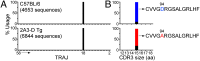
The self-reactivity of their T-cell antigen receptor (TCR) is thought to contribute to the development of immune regulatory cells, such as invariant NK T cells (iNKT). In the mouse, iNKT cells express TCRs composed of a unique Vα14-Jα18 rearrangement and recognize lipid antigens presented by CD1d molecules. We created mice expressing a transgenic TCR-β chain that confers high affinity for self-lipid/CD1d complexes when randomly paired with the mouse iNKT Vα14-Jα18 rearrangement to study their development. We show that although iNKT cells undergo agonist selection, their development is also shaped by negative selection in vivo. In addition, iNKT cells that avoid negative selection in these mice express natural sequence variants of the canonical TCR-α and decreased affinity for self/CD1d. However, limiting the affinity of the iNKT TCRs for "self" leads to inefficient Egr2 induction, poor expression of the iNKT lineage-specific zinc-finger transcription factor PLZF, inadequate proliferation of iNKT cell precursors, defects in trafficking, and impaired effector functions. Thus, proper development of fully functional iNKT cells is constrained by a limited range of TCR affinity that plays a key role in triggering the iNKT cell-differentiation pathway. These results provide a direct link between the affinity of the TCR expressed by T-cell precursors for self-antigens and the proper development of a unique population of lymphocytes essential to immune responses.
Keywords: deletion; thymus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed
-
- Kronenberg M. Toward an understanding of NKT cell biology: Progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. - PubMed
-
- Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13(2):101–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI018785/AI/NIAID NIH HHS/United States
- P30CA046934/CA/NCI NIH HHS/United States
- AI22295/AI/NIAID NIH HHS/United States
- AI18785/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- AI092108/AI/NIAID NIH HHS/United States
- T32 GM008497/GM/NIGMS NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- R37 AI018785/AI/NIAID NIH HHS/United States
- R01 AI092108/AI/NIAID NIH HHS/United States
- R56 AI018785/AI/NIAID NIH HHS/United States
- P01 AI022295/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

