Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin
- PMID: 24345744
- PMCID: PMC3870260
- DOI: 10.1128/mBio.00889-13
Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin
Abstract
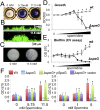
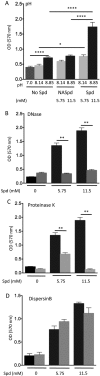
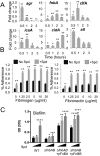
The arginine catabolic mobile element (ACME) is the largest genomic region distinguishing epidemic USA300 strains of methicillin-resistant Staphylococcus aureus (MRSA) from other S. aureus strains. However, the functional relevance of ACME to infection and disease has remained unclear. Using phylogenetic analysis, we have shown that the modular segments of ACME were assembled into a single genetic locus in Staphylococcus epidermidis and then horizontally transferred to the common ancestor of USA300 strains in an extremely recent event. Acquisition of one ACME gene, speG, allowed USA300 strains to withstand levels of polyamines (e.g., spermidine) produced in skin that are toxic to other closely related S. aureus strains. speG-mediated polyamine tolerance also enhanced biofilm formation, adherence to fibrinogen/fibronectin, and resistance to antibiotic and keratinocyte-mediated killing. We suggest that these properties gave USA300 a major selective advantage during skin infection and colonization, contributing to the extraordinary evolutionary success of this clone.
Importance: Over the past 15 years, methicillin-resistant Staphylococcus aureus (MRSA) has become a major public health problem. It is likely that adaptations in specific MRSA lineages (e.g., USA300) drove the spread of MRSA across the United States and allowed it to replace other, less-virulent S. aureus strains. We suggest that one major factor in the evolutionary success of MRSA may have been the acquisition of a gene (speG) that allows S. aureus to evade the toxicity of polyamines (e.g., spermidine and spermine) that are produced in human skin. Polyamine tolerance likely gave MRSA multiple fitness advantages, including the formation of more-robust biofilms, increased adherence to host tissues, and resistance to antibiotics and killing by human skin cells.
Figures



References
-
- Tenover FC, Goering RV. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64:441–446 - PubMed
-
- Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42:647–656 - PubMed
-
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771 - PubMed
-
- Otto M. 2011. A MRSA-terious enemy among us: end of the PVL controversy? Nat. Med. 17:169–170 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
