The reverse gyrase from Pyrobaculum calidifontis, a novel extremely thermophilic DNA topoisomerase endowed with DNA unwinding and annealing activities
- PMID: 24347172
- PMCID: PMC3916527
- DOI: 10.1074/jbc.M113.517649
The reverse gyrase from Pyrobaculum calidifontis, a novel extremely thermophilic DNA topoisomerase endowed with DNA unwinding and annealing activities
Abstract
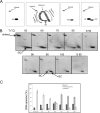
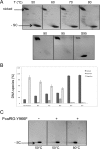
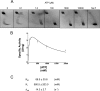
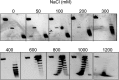
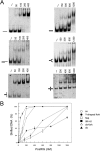
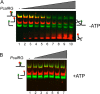
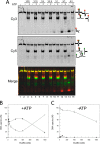
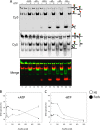
Reverse gyrase is a DNA topoisomerase specific for hyperthermophilic bacteria and archaea. It catalyzes the peculiar ATP-dependent DNA-positive supercoiling reaction and might be involved in the physiological adaptation to high growth temperature. Reverse gyrase comprises an N-terminal ATPase and a C-terminal topoisomerase domain, which cooperate in enzyme activity, but details of its mechanism of action are still not clear. We present here a functional characterization of PcalRG, a novel reverse gyrase from the archaeon Pyrobaculum calidifontis. PcalRG is the most robust and processive reverse gyrase known to date; it is active over a wide range of conditions, including temperature, ionic strength, and ATP concentration. Moreover, it holds a strong ATP-inhibited DNA cleavage activity. Most important, PcalRG is able to induce ATP-dependent unwinding of synthetic Holliday junctions and ATP-stimulated annealing of unconstrained single-stranded oligonucleotides. Combined DNA unwinding and annealing activities are typical of certain helicases, but until now were shown for no other reverse gyrase. Our results suggest for the first time that a reverse gyrase shares not only structural but also functional features with evolutionary conserved helicase-topoisomerase complexes involved in genome stability.
Keywords: DNA Enzymes; DNA Helicase; DNA Recombination; DNA Repair; DNA Topoisomerase; Holliday Junction; Thermophiles.
Figures
Similar articles
-
Synergic and opposing activities of thermophilic RecQ-like helicase and topoisomerase 3 proteins in Holliday junction processing and replication fork stabilization.J Biol Chem. 2012 Aug 31;287(36):30282-95. doi: 10.1074/jbc.M112.366377. Epub 2012 Jun 21. J Biol Chem. 2012. PMID: 22722926 Free PMC article.
-
Separate and combined biochemical activities of the subunits of a naturally split reverse gyrase.J Biol Chem. 2010 Dec 17;285(51):39637-45. doi: 10.1074/jbc.M110.173989. Epub 2010 Oct 6. J Biol Chem. 2010. PMID: 20929866 Free PMC article.
-
Reverse gyrase: an unusual DNA manipulator of hyperthermophilic organisms.Ital J Biochem. 2007 Jun;56(2):103-9. Ital J Biochem. 2007. PMID: 17722650 Review.
-
The archaeal topoisomerase reverse gyrase is a helix-destabilizing protein that unwinds four-way DNA junctions.J Biol Chem. 2010 Nov 19;285(47):36532-41. doi: 10.1074/jbc.M110.169029. Epub 2010 Sep 17. J Biol Chem. 2010. PMID: 20851892 Free PMC article.
-
Reverse gyrase and genome stability in hyperthermophilic organisms.Biochem Soc Trans. 2009 Feb;37(Pt 1):69-73. doi: 10.1042/BST0370069. Biochem Soc Trans. 2009. PMID: 19143604 Review.
Cited by
-
IND-enzymes: a repository for hydrolytic enzymes derived from thermophilic and psychrophilic bacterial species with potential industrial usage.Extremophiles. 2021 Jul;25(4):319-325. doi: 10.1007/s00792-021-01231-2. Epub 2021 May 7. Extremophiles. 2021. PMID: 33961119
-
Taxonomic and functional characterization of a microbial community from a volcanic englacial ecosystem in Deception Island, Antarctica.Sci Rep. 2019 Aug 21;9(1):12158. doi: 10.1038/s41598-019-47994-9. Sci Rep. 2019. PMID: 31434915 Free PMC article.
-
Genome stability: recent insights in the topoisomerase reverse gyrase and thermophilic DNA alkyltransferase.Extremophiles. 2014 Sep;18(5):895-904. doi: 10.1007/s00792-014-0662-9. Epub 2014 Aug 8. Extremophiles. 2014. PMID: 25102812 Review.
-
Recent advances in understanding extremophiles.F1000Res. 2019 Nov 13;8:F1000 Faculty Rev-1917. doi: 10.12688/f1000research.20765.1. eCollection 2019. F1000Res. 2019. PMID: 31781367 Free PMC article. Review.
-
Reverse gyrase--recent advances and current mechanistic understanding of positive DNA supercoiling.Nucleic Acids Res. 2014 Jul;42(13):8200-13. doi: 10.1093/nar/gku589. Epub 2014 Jul 10. Nucleic Acids Res. 2014. PMID: 25013168 Free PMC article. Review.
References
-
- Nadal M. (2007) Reverse gyrase: an insight into the role of DNA-topoisomerases. Biochimie 89, 447–455 - PubMed
-
- Perugino G., Valenti A., D'amaro A., Rossi M., Ciaramella M. (2009) Reverse gyrase and genome stability in hyperthermophilic organisms. Biochem. Soc. Trans. 37, 69–73 - PubMed
-
- Heine M., Chandra S. B. (2009) The linkage between reverse gyrase and hyperthermophiles: a review of their invariable association. J. Microbiol. 47, 229–234 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

