A comprehensive phylogenetic analysis of deadenylases
- PMID: 24348009
- PMCID: PMC3859875
- DOI: 10.4137/EBO.S12746
A comprehensive phylogenetic analysis of deadenylases
Abstract
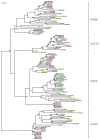
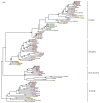
Deadenylases catalyze the shortening of the poly(A) tail at the messenger ribonucleic acid (mRNA) 3'-end in eukaryotes. Therefore, these enzymes influence mRNA decay, and constitute a major emerging group of promising anti-cancer pharmacological targets. Herein, we conducted full phylogenetic analyses of the deadenylase homologs in all available genomes in an effort to investigate evolutionary relationships between the deadenylase families and to identify invariant residues, which probably play key roles in the function of deadenylation across species. Our study includes both major Asp-Glu-Asp-Asp (DEDD) and exonuclease-endonuclease-phospatase (EEP) deadenylase superfamilies. The phylogenetic analysis has provided us with important information regarding conserved and invariant deadenylase amino acids across species. Knowledge of the phylogenetic properties and evolution of the domain of deadenylases provides the foundation for the targeted drug design in the pharmaceutical industry and modern exonuclease anti-cancer scientific research.
Keywords: deadenylases; evolution; molecular modelling; phylogenesis.
Figures

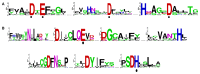
Similar articles
-
Saccharomyces cerevisiae Ngl3p is an active 3'-5' exonuclease with a specificity towards poly-A RNA reminiscent of cellular deadenylases.Nucleic Acids Res. 2012 Jan;40(2):837-46. doi: 10.1093/nar/gkr782. Epub 2011 Sep 29. Nucleic Acids Res. 2012. PMID: 21965533 Free PMC article.
-
Heterogeneity and complexity within the nuclease module of the Ccr4-Not complex.Front Genet. 2013 Dec 23;4:296. doi: 10.3389/fgene.2013.00296. Front Genet. 2013. PMID: 24391663 Free PMC article. Review.
-
Biochemical and in silico identification of the active site and the catalytic mechanism of the circadian deadenylase HESPERIN.FEBS Open Bio. 2022 May;12(5):1036-1049. doi: 10.1002/2211-5463.13011. Epub 2022 Mar 29. FEBS Open Bio. 2022. PMID: 33095977 Free PMC article.
-
Deadenylation: enzymes, regulation, and functional implications.Wiley Interdiscip Rev RNA. 2014 May-Jun;5(3):421-43. doi: 10.1002/wrna.1221. Epub 2014 Feb 12. Wiley Interdiscip Rev RNA. 2014. PMID: 24523229 Review.
-
The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans.J Cell Sci. 2013 Sep 15;126(Pt 18):4274-85. doi: 10.1242/jcs.132936. Epub 2013 Jul 10. J Cell Sci. 2013. PMID: 23843623
Cited by
-
Temperature-dependent fasciation mutants provide a link between mitochondrial RNA processing and lateral root morphogenesis.Elife. 2021 Jan 14;10:e61611. doi: 10.7554/eLife.61611. Elife. 2021. PMID: 33443014 Free PMC article.
-
A unique system for regulating mitochondrial mRNA poly(A) status and stability in plants.Plant Signal Behav. 2014;9(10):e973809. doi: 10.4161/15592324.2014.973809. Plant Signal Behav. 2014. PMID: 25482772 Free PMC article.
-
Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development.Nucleic Acids Res. 2016 Oct 14;44(18):8908-8920. doi: 10.1093/nar/gkw709. Epub 2016 Aug 11. Nucleic Acids Res. 2016. PMID: 27515512 Free PMC article.
-
Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems.RNA Biol. 2014;11(9):1122-36. doi: 10.4161/rna.34406. RNA Biol. 2014. PMID: 25483043 Free PMC article. Review.
-
TOE1 acts as a 3' exonuclease for telomerase RNA and regulates telomere maintenance.Nucleic Acids Res. 2019 Jan 10;47(1):391-405. doi: 10.1093/nar/gky1019. Nucleic Acids Res. 2019. PMID: 30371886 Free PMC article.
References
-
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–26. - PubMed
-
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104(3):377–86. - PubMed
-
- Uchida N, Hoshino S, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem. 2004;279(2):1383–91. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

