Evaluating the potential of poly(beta-amino ester) nanoparticles for reprogramming human fibroblasts to become induced pluripotent stem cells
- PMID: 24348039
- PMCID: PMC3857166
- DOI: 10.2147/IJN.S53830
Evaluating the potential of poly(beta-amino ester) nanoparticles for reprogramming human fibroblasts to become induced pluripotent stem cells
Abstract
Background: Gene delivery can potentially be used as a therapeutic for treating genetic diseases, including neurodegenerative diseases, as well as an enabling technology for regenerative medicine. A central challenge in many gene delivery applications is having a safe and effective delivery method. We evaluated the use of a biodegradable poly(beta-amino ester) nanoparticle-based nonviral protocol and compared this with an electroporation-based approach to deliver episomal plasmids encoding reprogramming factors for generation of human induced pluripotent stem cells (hiPSCs) from human fibroblasts.
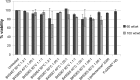
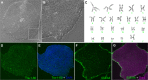
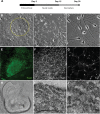
Methods: A polymer library was screened to identify the polymers most promising for gene delivery to human fibroblasts. Feeder-independent culturing protocols were developed for nanoparticle-based and electroporation-based reprogramming. The cells reprogrammed by both polymeric nanoparticle-based and electroporation-based nonviral methods were characterized by analysis of pluripotency markers and karyotypic stability. The hiPSC-like cells were further differentiated toward the neural lineage to test their potential for neurodegenerative retinal disease modeling.
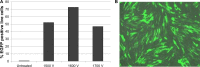
Results: 1-(3-aminopropyl)-4-methylpiperazine end-terminated poly(1,4-butanediol diacry-late-co-4-amino-1-butanol) polymer (B4S4E7) self-assembled with plasmid DNA to form nanoparticles that were more effective than leading commercially available reagents, including Lipofectamine® 2000, FuGENE® HD, and 25 kDa branched polyethylenimine, for nonviral gene transfer. B4S4E7 nanoparticles showed effective gene delivery to IMR-90 human primary fibroblasts and to dermal fibroblasts derived from a patient with retinitis pigmentosa, and enabled coexpression of exogenously delivered genes, as is needed for reprogramming. The karyotypically normal hiPSC-like cells generated by conventional electroporation, but not by poly(beta-amino ester) reprogramming, could be differentiated toward the neuronal lineage, specifically pseudostratified optic cups.
Conclusion: This study shows that certain nonviral reprogramming methods may not necessarily be safer than viral approaches and that maximizing exogenous gene expression of reprogramming factors is not sufficient to ensure successful reprogramming.
Keywords: human fibroblasts; induced pluripotent stem cells; poly(beta-amino ester) nanoparticles; reprogramming.
Figures








References
-
- Jin ZB, Okamoto S, Mandai M, Takahashi M. Induced pluripotent stem cells for retinal degenerative diseases: a new perspective on the challenges. J Genet. 2009;88(4):417–424. - PubMed
-
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. - PubMed
-
- Verma IM, Somia N. Gene therapy – promises, problems and prospects. Nature. 1997;389(6648):239–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

