Next-generation sequence assembly: four stages of data processing and computational challenges
- PMID: 24348224
- PMCID: PMC3861042
- DOI: 10.1371/journal.pcbi.1003345
Next-generation sequence assembly: four stages of data processing and computational challenges
Abstract
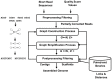
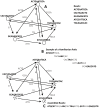
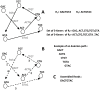
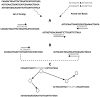
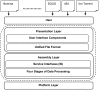
Decoding DNA symbols using next-generation sequencers was a major breakthrough in genomic research. Despite the many advantages of next-generation sequencers, e.g., the high-throughput sequencing rate and relatively low cost of sequencing, the assembly of the reads produced by these sequencers still remains a major challenge. In this review, we address the basic framework of next-generation genome sequence assemblers, which comprises four basic stages: preprocessing filtering, a graph construction process, a graph simplification process, and postprocessing filtering. Here we discuss them as a framework of four stages for data analysis and processing and survey variety of techniques, algorithms, and software tools used during each stage. We also discuss the challenges that face current assemblers in the next-generation environment to determine the current state-of-the-art. We recommend a layered architecture approach for constructing a general assembler that can handle the sequences generated by different sequencing platforms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Voelkerding KV, Dames SA, Durtschi JD (2009) Next-generation sequencing: from basic research to diagnostics. Clin Chem 55: 641–658. - PubMed
-
- Helmy M, Sugiyama N, Tomita M, Ishihama Y (2012) Mass spectrum sequential subtraction speeds up searching large peptide MS/MS spectra datasets against large nucleotide databases for proteogenomics. Genes Cells 17: 633–644. - PubMed
-
- Helmy M, Tomita M, Ishihama Y (2011) Peptide identification by searching large-scale tandem mass spectra against large databases: bioinformatics methods in proteogenomics. Genes, Genomes and Genomics 6: 76–85.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

