The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana
- PMID: 24348487
- PMCID: PMC3842039
- DOI: 10.3389/fpls.2013.00480
The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana
Abstract
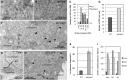
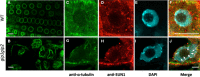
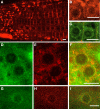
During interphase, the microtubular cytoskeleton of cycling plant cells is organized in both cortical and perinuclear arrays. Perinuclear microtubules (MTs) are nucleated from γ-Tubulin Complexes (γ-TuCs) located at the surface of the nucleus. The molecular mechanisms of γ-TuC association to the nuclear envelope (NE) are currently unknown. The γ-TuC Protein 3 (GCP3)-Interacting Protein 1 (GIP1) is the smallest γ-TuC component identified so far. AtGIP1 and its homologous protein AtGIP2 participate in the localization of active γ-TuCs at interphasic and mitotic MT nucleation sites. Arabidopsis gip1gip2 mutants are impaired in establishing a fully functional mitotic spindle and exhibit severe developmental defects. In this study, gip1gip2 knock down mutants were further characterized at the cellular level. In addition to defects in both the localization of γ-TuC core proteins and MT fiber robustness, gip1gip2 mutants exhibited a severe alteration of the nuclear shape associated with an abnormal distribution of the nuclear pore complexes. Simultaneously, they showed a misorganization of the inner nuclear membrane protein AtSUN1. Furthermore, AtGIP1 was identified as an interacting partner of AtTSA1 which was detected, like the AtGIP proteins, at the NE. These results provide the first evidence for the involvement of a γ-TuC component in both nuclear shaping and NE organization. Functional hypotheses are discussed in order to propose a model for a GIP-dependent nucleo-cytoplasmic continuum.
Keywords: Arabidopsis thaliana; AtGIP1/MOZART1; AtTSA1; gamma-tubulin complex; nuclear envelope.
Figures




References
-
- Bajer A., Mole-Bajer J. (1969). Formation of spindle fibers, kinetochore orientation, and behavior of the nuclear envelope during mitosis in endosperm. Chromosoma 27, 448–484 10.1007/BF00325682 - DOI
-
- Ciska M., Moreno Diaz de la Espina S. (2013). NMCP/LINC proteins: putative lamin analogs in plants? Plant signal. Behav. 8 Available online at: http://www.ncbi.nlm.nih.gov/pubmed/24128696 10.4161/psb.26669 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

