Desiccation tolerance in resurrection plants: new insights from transcriptome, proteome and metabolome analysis
- PMID: 24348488
- PMCID: PMC3842845
- DOI: 10.3389/fpls.2013.00482
Desiccation tolerance in resurrection plants: new insights from transcriptome, proteome and metabolome analysis
Abstract
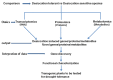
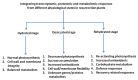
Most higher plants are unable to survive desiccation to an air-dried state. An exception is a small group of vascular angiosperm plants, termed resurrection plants. They have evolved unique mechanisms of desiccation tolerance and thus can tolerate severe water loss, and mostly adjust their water content with the relative humidity in the environment. Desiccation tolerance is a complex phenomenon and depends on the regulated expression of numerous genes during dehydration and subsequent rehydration. Most of the resurrection plants have a large genome and are difficult to transform which makes them unsuitable for genetic approaches. However, technical advances have made it possible to analyze changes in gene expression on a large-scale. These approaches together with comparative studies with non-desiccation tolerant plants provide novel insights into the molecular processes required for desiccation tolerance and will shed light on identification of orphan genes with unknown functions. Here, we review large-scale recent transcriptomic, proteomic, and metabolomic studies that have been performed in desiccation tolerant plants and discuss how these studies contribute to understanding the molecular basis of desiccation tolerance.
Keywords: desiccation tolerance; metabolomics; proteomics; resurrection plants; transcriptomics.
Figures
References
-
- Alamillo J. M., Bartels D. (1996). Light and stage of development influence the expression of desiccation-induced genes in the resurrection plant Craterostigma plantagineum. Plant Cell Environ. 19 300–310 10.1111/j.1365-3040.1996.tb00252.x - DOI
-
- Alpert P., Oliver M. (2002). “Drying without dying,” in Desiccation and Survival in Plants: Drying without Dying eds Black M., Pritchard H. (New York, NY: CABI Publishing; ) 1–43
LinkOut - more resources
Full Text Sources
Other Literature Sources

