On protocols and measures for the validation of supervised methods for the inference of biological networks
- PMID: 24348517
- PMCID: PMC3848415
- DOI: 10.3389/fgene.2013.00262
On protocols and measures for the validation of supervised methods for the inference of biological networks
Abstract
Networks provide a natural representation of molecular biology knowledge, in particular to model relationships between biological entities such as genes, proteins, drugs, or diseases. Because of the effort, the cost, or the lack of the experiments necessary for the elucidation of these networks, computational approaches for network inference have been frequently investigated in the literature. In this paper, we examine the assessment of supervised network inference. Supervised inference is based on machine learning techniques that infer the network from a training sample of known interacting and possibly non-interacting entities and additional measurement data. While these methods are very effective, their reliable validation in silico poses a challenge, since both prediction and validation need to be performed on the basis of the same partially known network. Cross-validation techniques need to be specifically adapted to classification problems on pairs of objects. We perform a critical review and assessment of protocols and measures proposed in the literature and derive specific guidelines how to best exploit and evaluate machine learning techniques for network inference. Through theoretical considerations and in silico experiments, we analyze in depth how important factors influence the outcome of performance estimation. These factors include the amount of information available for the interacting entities, the sparsity and topology of biological networks, and the lack of experimentally verified non-interacting pairs.
Keywords: ROC curves; biological network inference; cross-validation; evaluation protocols; precision-recall curves; supervised learning.
Figures
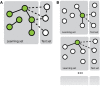

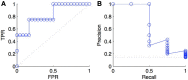

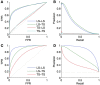
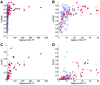
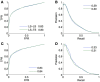
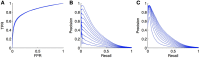
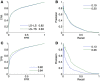
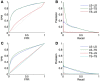
References
-
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32 10.1023/A:1017934522171 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

