Discovering epistasis in large scale genetic association studies by exploiting graphics cards
- PMID: 24348518
- PMCID: PMC3848199
- DOI: 10.3389/fgene.2013.00266
Discovering epistasis in large scale genetic association studies by exploiting graphics cards
Abstract
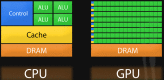
Despite the enormous investments made in collecting DNA samples and generating germline variation data across thousands of individuals in modern genome-wide association studies (GWAS), progress has been frustratingly slow in explaining much of the heritability in common disease. Today's paradigm of testing independent hypotheses on each single nucleotide polymorphism (SNP) marker is unlikely to adequately reflect the complex biological processes in disease risk. Alternatively, modeling risk as an ensemble of SNPs that act in concert in a pathway, and/or interact non-additively on log risk for example, may be a more sensible way to approach gene mapping in modern studies. Implementing such analyzes genome-wide can quickly become intractable due to the fact that even modest size SNP panels on modern genotype arrays (500k markers) pose a combinatorial nightmare, require tens of billions of models to be tested for evidence of interaction. In this article, we provide an in-depth analysis of programs that have been developed to explicitly overcome these enormous computational barriers through the use of processors on graphics cards known as Graphics Processing Units (GPU). We include tutorials on GPU technology, which will convey why they are growing in appeal with today's numerical scientists. One obvious advantage is the impressive density of microprocessor cores that are available on only a single GPU. Whereas high end servers feature up to 24 Intel or AMD CPU cores, the latest GPU offerings from nVidia feature over 2600 cores. Each compute node may be outfitted with up to 4 GPU devices. Success on GPUs varies across problems. However, epistasis screens fare well due to the high degree of parallelism exposed in these problems. Papers that we review routinely report GPU speedups of over two orders of magnitude (>100x) over standard CPU implementations.
Keywords: CUDA tutorial; GPU programming; epistasis; gene–gene interactions; high performance computing.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

