Laboratory-based prospective surveillance for community outbreaks of Shigella spp. in Argentina
- PMID: 24349586
- PMCID: PMC3861122
- DOI: 10.1371/journal.pntd.0002521
Laboratory-based prospective surveillance for community outbreaks of Shigella spp. in Argentina
Abstract
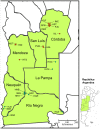
Background: To implement effective control measures, timely outbreak detection is essential. Shigella is the most common cause of bacterial diarrhea in Argentina. Highly resistant clones of Shigella have emerged, and outbreaks have been recognized in closed settings and in whole communities. We hereby report our experience with an evolving, integrated, laboratory-based, near real-time surveillance system operating in six contiguous provinces of Argentina during April 2009 to March 2012.
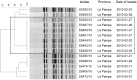
Methodology: To detect localized shigellosis outbreaks timely, we used the prospective space-time permutation scan statistic algorithm of SaTScan, embedded in WHONET software. Twenty three laboratories sent updated Shigella data on a weekly basis to the National Reference Laboratory. Cluster detection analysis was performed at several taxonomic levels: for all Shigella spp., for serotypes within species and for antimicrobial resistance phenotypes within species. Shigella isolates associated with statistically significant signals (clusters in time/space with recurrence interval ≥365 days) were subtyped by pulsed field gel electrophoresis (PFGE) using PulseNet protocols.
Principal findings: In three years of active surveillance, our system detected 32 statistically significant events, 26 of them identified before hospital staff was aware of any unexpected increase in the number of Shigella isolates. Twenty-six signals were investigated by PFGE, which confirmed a close relationship among the isolates for 22 events (84.6%). Seven events were investigated epidemiologically, which revealed links among the patients. Seventeen events were found at the resistance profile level. The system detected events of public health importance: infrequent resistance profiles, long-lasting and/or re-emergent clusters and events important for their duration or size, which were reported to local public health authorities.
Conclusions/significance: The WHONET-SaTScan system may serve as a model for surveillance and can be applied to other pathogens, implemented by other networks, and scaled up to national and international levels for early detection and control of outbreaks.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Automated use of WHONET and SaTScan to detect outbreaks of Shigella spp. using antimicrobial resistance phenotypes.Epidemiol Infect. 2010 Jun;138(6):873-83. doi: 10.1017/S0950268809990884. Epub 2009 Oct 2. Epidemiol Infect. 2010. PMID: 19796449 Free PMC article.
-
A unified approach to molecular epidemiology investigations: tools and patterns in California as a case study for endemic shigellosis.BMC Infect Dis. 2009 Nov 24;9:184. doi: 10.1186/1471-2334-9-184. BMC Infect Dis. 2009. PMID: 19930709 Free PMC article.
-
Facilitated molecular typing of Shigella isolates using ERIC-PCR.Am J Trop Med Hyg. 2012 Jun;86(6):1018-25. doi: 10.4269/ajtmh.2012.11-0671. Am J Trop Med Hyg. 2012. PMID: 22665611 Free PMC article.
-
Short report: analysis of clonal relationship among Shigella sonnei isolates circulating in Argentina.Epidemiol Infect. 2007 May;135(4):681-7. doi: 10.1017/S0950268806007230. Epub 2006 Sep 26. Epidemiol Infect. 2007. PMID: 16999876 Free PMC article. Review.
-
PulseNet China, a model for future laboratory-based bacterial infectious disease surveillance in China.Front Med. 2012 Dec;6(4):366-75. doi: 10.1007/s11684-012-0214-6. Epub 2012 Nov 3. Front Med. 2012. PMID: 23124882 Review.
Cited by
-
A cohort study to define the age-specific incidence and risk factors of Shigella diarrhoeal infections in Vietnamese children: a study protocol.BMC Public Health. 2014 Dec 17;14:1289. doi: 10.1186/1471-2458-14-1289. BMC Public Health. 2014. PMID: 25514820 Free PMC article.
-
Surveillance of antimicrobial resistance and evolving microbial populations in Vermont: 2011-2018.Expert Rev Anti Infect Ther. 2020 Oct;18(10):1055-1062. doi: 10.1080/14787210.2020.1776114. Epub 2020 Jun 18. Expert Rev Anti Infect Ther. 2020. PMID: 32552054 Free PMC article.
-
Web-Based Apps for Responding to Acute Infectious Disease Outbreaks in the Community: Systematic Review.JMIR Public Health Surveill. 2021 Apr 21;7(4):e24330. doi: 10.2196/24330. JMIR Public Health Surveill. 2021. PMID: 33881406 Free PMC article.
-
Prospective Spatiotemporal Cluster Detection Using SaTScan: Tutorial for Designing and Fine-Tuning a System to Detect Reportable Communicable Disease Outbreaks.JMIR Public Health Surveill. 2024 Jun 11;10:e50653. doi: 10.2196/50653. JMIR Public Health Surveill. 2024. PMID: 38861711 Free PMC article.
-
Online platform for applying space-time scan statistics for prospectively detecting emerging hot spots of dengue fever.Int J Health Geogr. 2016 Nov 25;15(1):43. doi: 10.1186/s12942-016-0072-6. Int J Health Geogr. 2016. PMID: 27884135 Free PMC article.
References
-
- WHO Global Foodborne Infections Network (GFN). Data and statistics. Available at: http://www.who.int/gfn/en/. Accessed 8 October 2012.
-
- WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). Antimicrobial Resistance Available at:http://www.who.int/foodborne_disease/resistance/agisar/en/. Accessed 8 October 2012
-
- Swaminathan B, Gerner-Smidt P, Ng LK, Lukinmaa S, Kam KM, et al. (2006) Building PulseNet International: an interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathog Dis Spring 3 (1) 36–50. - PubMed
-
- O'Brien TF, Stelling J (2011) Integrated multilevel surveillance of the world's infecting microbes and their resistance to antimicrobial agents. Clin Microbiol Rev Apr; 24 (2) 281–95 doi: 10.1128/CMR.00021-10 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

