Biased gene expression in early honeybee larval development
- PMID: 24350621
- PMCID: PMC3878232
- DOI: 10.1186/1471-2164-14-903
Biased gene expression in early honeybee larval development
Abstract
Background: Female larvae of the honeybee (Apis mellifera) develop into either queens or workers depending on nutrition. This nutritional stimulus triggers different developmental trajectories, resulting in adults that differ from each other in physiology, behaviour and life span.
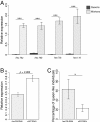
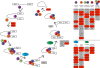
Results: To understand how these trajectories are established we have generated a comprehensive atlas of gene expression throughout larval development. We found substantial differences in gene expression between worker and queen-destined larvae at 6 hours after hatching. Some of these early changes in gene expression are maintained throughout larval development, indicating that caste-specific developmental trajectories are established much earlier than previously thought. Within our gene expression data we identified processes that potentially underlie caste differentiation. Queen-destined larvae have higher expression of genes involved in transcription, translation and protein folding early in development with a later switch to genes involved in energy generation. Using RNA interference, we were able to demonstrate that one of these genes, hexamerin 70b, has a role in caste differentiation. Both queen and worker developmental trajectories are associated with the expression of genes that have alternative splice variants, although only a single variant of a gene tends to be differentially expressed in a given caste.
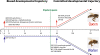
Conclusions: Our data, based on the biases in gene expression early in development together with published data, supports the idea that caste development in the honeybee consists of two phases; an initial biased phase of development, where larvae can still switch to the other caste by differential feeding, followed by commitment to a particular developmental trajectory.
Figures
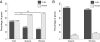

References
-
- Huber F. New observations on the natural history of bees. London: Longman, Hurst, Rees and Orme; 1821.
-
- Dixon SE, Shuel RW. Studies in the mode of action of royal jelly in honeybee development III. The effect of experimental variation in diet on growth and metabolism of honeybee larvae. Can J Zool. 1963;14:733–739. doi: 10.1139/z63-045. - DOI
-
- Haydak MH. Honey bee nutrition. Annu Rev Entomol. 1970;14:143–156. doi: 10.1146/annurev.en.15.010170.001043. - DOI
-
- Shuel R, Dixon S. The early establishment of dimorphism in the female honeybee, Apis mellifera L. Insect Soc. 1960;14(3):265–282. doi: 10.1007/BF02224497. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

