Combining single RNA sensitive probes with subdiffraction-limited and live-cell imaging enables the characterization of virus dynamics in cells
- PMID: 24351207
- PMCID: PMC3906890
- DOI: 10.1021/nn405998v
Combining single RNA sensitive probes with subdiffraction-limited and live-cell imaging enables the characterization of virus dynamics in cells
Abstract
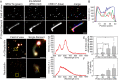
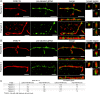
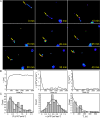
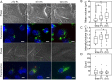
The creation of fluorescently labeled viruses is currently limited by the length of imaging observation time (e.g., labeling an envelope protein) and the rescue of viral infectivity (e.g., encoding a GFP protein). Using single molecule sensitive RNA hybridization probes delivered to the cytoplasm of infected cells, we were able to isolate individual, infectious, fluorescently labeled human respiratory syncytial virus virions. This was achieved without affecting viral mRNA expression, viral protein expression, or infectivity. Measurements included the characterization of viral proteins and genomic RNA in a single virion using dSTORM, the development of a GFP fusion assay, and the development of a pulse-chase assay for viral RNA production that allowed for the detection of both initial viral RNA and nascent RNA production at designated times postinfection. Live-cell measurements included imaging and characterization of filamentous virion fusion and the quantification of virus replication within the same cell over an eight-hour period. Using probe-labeled viruses, individual viral particles can be characterized at subdiffraction-limited resolution, and viral infections can be quantified in single cells over an entire cycle of replication. The implication of this development is that MTRIP labeling of viral RNA during virus assembly has the potential to become a general methodology for the labeling and study of many important RNA viruses.
Figures


Similar articles
-
Imaging viral RNA using multiply labeled tetravalent RNA imaging probes in live cells.Methods. 2016 Apr 1;98:91-98. doi: 10.1016/j.ymeth.2016.02.006. Epub 2016 Feb 12. Methods. 2016. PMID: 26875782 Free PMC article. Review.
-
Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene.J Virol. 2001 Aug;75(15):6825-34. doi: 10.1128/JVI.75.15.6825-6834.2001. J Virol. 2001. PMID: 11435561 Free PMC article.
-
Chimeric bovine respiratory syncytial virus with glycoprotein gene substitutions from human respiratory syncytial virus (HRSV): effects on host range and evaluation as a live-attenuated HRSV vaccine.J Virol. 2000 Feb;74(3):1187-99. doi: 10.1128/jvi.74.3.1187-1199.2000. J Virol. 2000. PMID: 10627529 Free PMC article.
-
Single molecule sensitive multivalent polyethylene glycol probes for RNA imaging.Bioconjug Chem. 2010 Mar 17;21(3):483-8. doi: 10.1021/bc9003876. Epub 2010 Feb 8. Bioconjug Chem. 2010. PMID: 20141184
-
Molecular mechanisms driving respiratory syncytial virus assembly.Future Microbiol. 2013 Jan;8(1):123-31. doi: 10.2217/fmb.12.132. Future Microbiol. 2013. PMID: 23252497 Free PMC article. Review.
Cited by
-
Regeneration of infarcted mouse hearts by cardiovascular tissue formed via the direct reprogramming of mouse fibroblasts.Nat Biomed Eng. 2021 Aug;5(8):880-896. doi: 10.1038/s41551-021-00783-0. Epub 2021 Aug 23. Nat Biomed Eng. 2021. PMID: 34426676 Free PMC article.
-
Exploring the secrets of virus entry: the first respiratory syncytial virus carrying beta lactamase.Front Microbiol. 2024 Feb 16;15:1339569. doi: 10.3389/fmicb.2024.1339569. eCollection 2024. Front Microbiol. 2024. PMID: 38455070 Free PMC article.
-
Actin cytoskeleton remodeling disrupts physical barriers to infection and presents entry receptors to respiratory syncytial virus.J Gen Virol. 2023 Nov;104(11):001923. doi: 10.1099/jgv.0.001923. J Gen Virol. 2023. PMID: 38015055 Free PMC article.
-
Mirror-enhanced super-resolution microscopy.Light Sci Appl. 2016;5(6):e16134-. doi: 10.1038/lsa.2016.134. Epub 2016 Jun 17. Light Sci Appl. 2016. PMID: 27398242 Free PMC article.
-
Characterizing exogenous mRNA delivery, trafficking, cytoplasmic release and RNA-protein correlations at the level of single cells.Nucleic Acids Res. 2017 Jul 7;45(12):e113. doi: 10.1093/nar/gkx290. Nucleic Acids Res. 2017. PMID: 28449134 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

