Towards lipidomics of low-abundant species for exploring tumor heterogeneity guided by high-resolution mass spectrometry imaging
- PMID: 24351834
- PMCID: PMC3876128
- DOI: 10.3390/ijms141224560
Towards lipidomics of low-abundant species for exploring tumor heterogeneity guided by high-resolution mass spectrometry imaging
Abstract
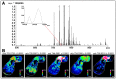
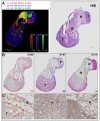
Many studies have evidenced the main role of lipids in physiological and also pathological processes such as cancer, diabetes or neurodegenerative diseases. The identification and the in situ localization of specific low-abundant lipid species involved in cancer biology are still challenging for both fundamental studies and lipid marker discovery. In this paper, we report the identification and the localization of specific isobaric minor phospholipids in human breast cancer xenografts by FTICR MALDI imaging supported by histochemistry. These potential candidates can be further confirmed by liquid chromatography coupled with electrospray mass spectrometry (LC-ESI-MS) after extraction from the region of interest defined by MALDI imaging. Finally, this study highlights the importance of characterizing the heterogeneous distribution of low-abundant lipid species, relevant in complex histological samples for biological purposes.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

