Functional anatomy of an allosteric protein
- PMID: 24352193
- PMCID: PMC4407639
- DOI: 10.1038/ncomms3984
Functional anatomy of an allosteric protein
Abstract
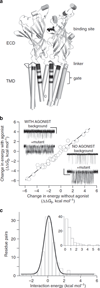
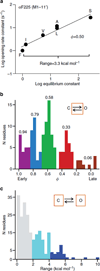
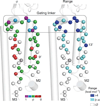
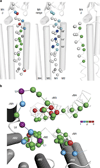
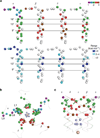
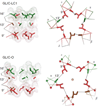
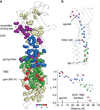
Synaptic receptors are allosteric proteins that switch on and off to regulate cell signalling. Here, we use single-channel electrophysiology to measure and map energy changes in the gating conformational change of a nicotinic acetylcholine receptor. Two separated regions in the α-subunits--the transmitter-binding sites and αM2-αM3 linkers in the membrane domain--have the highest ϕ-values (change conformation the earliest), followed by the extracellular domain, most of the membrane domain and the gate. Large gating-energy changes occur at the transmitter-binding sites, α-subunit interfaces, the αM1 helix and the gate. We hypothesize that rearrangements of the linkers trigger the global allosteric transition, and that the hydrophobic gate unlocks in three steps. The mostly local character of side-chain energy changes and the similarly high ϕ-values of separated domains, both with and without ligands, suggest that gating is not strictly a mechanical process initiated by the affinity change for the agonist.
Figures


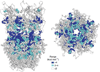
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

