Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies
- PMID: 24352443
- PMCID: PMC3958090
- DOI: 10.1128/JVI.02853-13
Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies
Abstract
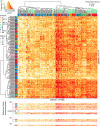
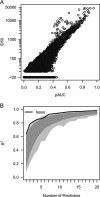
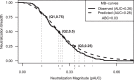
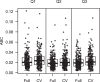
Standardized assessments of HIV-1 vaccine-elicited neutralizing antibody responses are complicated by the genetic and antigenic variability of the viral envelope glycoproteins (Envs). To address these issues, suitable reference strains are needed that are representative of the global epidemic. Several panels have been recommended previously, but no clear answers have been available on how many and which strains are best suited for this purpose. We used a statistical model selection method to identify a global panel of reference Env clones from among 219 Env-pseudotyped viruses assayed in TZM-bl cells with sera from 205 HIV-1-infected individuals. The Envs and sera were sampled globally from diverse geographic locations and represented all major genetic subtypes and circulating recombinant forms of the virus. Assays with a panel size of only nine viruses adequately represented the spectrum of HIV-1 serum neutralizing activity seen with the larger panel of 219 viruses. An optimal panel of nine viruses was selected and augmented with three additional viruses for greater genetic and antigenic coverage. The spectrum of HIV-1 serum neutralizing activity seen with the final 12-virus panel closely approximated the activity seen with subtype-matched viruses. Moreover, the final panel was highly sensitive for detection of many of the known broadly neutralizing antibodies. For broader assay applications, all 12 Env clones were converted to infectious molecular clones using a proviral backbone carrying a Renilla luciferase reporter gene (Env.IMC.LucR viruses). This global panel should facilitate highly standardized assessments of vaccine-elicited neutralizing antibodies across multiple HIV-1 vaccine platforms in different parts of the world.
Importance: An effective HIV-1 vaccine will need to overcome the extraordinary genetic variability of the virus, where most variation occurs in the viral envelope glycoproteins that are the sole targets for neutralizing antibodies. Efforts to elicit broadly cross-reactive neutralizing antibodies that will protect against infection by most circulating strains of the virus are guided in part by in vitro assays that determine the ability of vaccine-elicited antibodies to neutralize genetically diverse HIV-1 variants. Until now, little information was available on how many and which strains of the virus are best suited for this purpose. We applied robust statistical methods to evaluate a large neutralization data set and identified a small panel of viruses that are a good representation of the global epidemic. The neutralization properties of this new panel of reference strains should facilitate the development of an effective HIV-1 vaccine.
Figures








References
-
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessments of neutralizing antibodies. J. Virol. 84:1439–1452. 10.1128/JVI.02108-09 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

