Nucleic acid chaperone activity associated with the arginine-rich domain of human hepatitis B virus core protein
- PMID: 24352445
- PMCID: PMC3958103
- DOI: 10.1128/JVI.03235-13
Nucleic acid chaperone activity associated with the arginine-rich domain of human hepatitis B virus core protein
Abstract
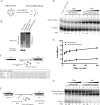
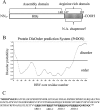
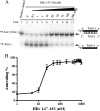
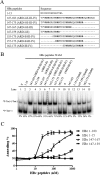
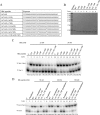
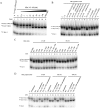
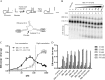
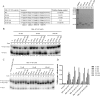
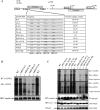
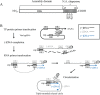
Hepatitis B virus (HBV) DNA replication occurs within the HBV icosahedral core particles. HBV core protein (HBc) contains an arginine-rich domain (ARD) at its carboxyl terminus. This ARD domain of HBc 149-183 is known to be important for viral replication but not known to have a structure. Recently, nucleocapsid proteins of several viruses have been shown to contain nucleic acid chaperone activity, which can facilitate structural rearrangement of viral genome. Major features of nucleic acid chaperones include highly basic amino acid residues and flexible protein structure. To test the nucleic acid chaperone hypothesis for HBc ARD, we first used the disassembled full-length HBc from Escherichia coli to analyze the nucleic acid annealing and strand displacement activities. To exclude the potential contamination of chaperones from E. coli, we designed synthetic HBc ARD peptides with different lengths and serine phosphorylations. We demonstrated that HBc ARD peptide can behave like a bona fide nucleic acid chaperone and that the chaperone activity depends on basic residues of the ARD domain. The loss of chaperone activity by arginine-to-alanine substitutions in the ARD can be rescued by restoring basic residues in the ARD. Furthermore, the chaperone activity is subject to regulation by phosphorylation and dephosphorylation at the HBc ARD. Interestingly, the HBc ARD can enhance in vitro cleavage activity of RNA substrate by a hammerhead ribozyme. We discuss here the potential significance of the HBc ARD chaperone activity in the context of viral DNA replication, in particular, at the steps of primer translocations and circularization of linear replicative intermediates.
Importance: Hepatitis B virus is a major human pathogen. At present, no effective treatment can completely eradicate the virus from patients with chronic hepatitis B. We report here a novel chaperone activity associated with the viral core protein. Our discovery could lead to a new drug design for more effective treatment against hepatitis B virus in the future.
Figures
Similar articles
-
Testing the balanced electrostatic interaction hypothesis of hepatitis B virus DNA synthesis by using an in vivo charge rebalance approach.J Virol. 2010 Mar;84(5):2340-51. doi: 10.1128/JVI.01666-09. Epub 2009 Dec 16. J Virol. 2010. PMID: 20015989 Free PMC article.
-
Conserved Lysine Residues of Hepatitis B Virus Core Protein Are Not Required for Covalently Closed Circular DNA Formation.J Virol. 2022 Aug 10;96(15):e0071822. doi: 10.1128/jvi.00718-22. Epub 2022 Jul 18. J Virol. 2022. PMID: 35867543 Free PMC article.
-
Nuclear export and import of human hepatitis B virus capsid protein and particles.PLoS Pathog. 2010 Oct 28;6(10):e1001162. doi: 10.1371/journal.ppat.1001162. PLoS Pathog. 2010. PMID: 21060813 Free PMC article.
-
Phosphorylation of the Arginine-Rich C-Terminal Domains of the Hepatitis B Virus (HBV) Core Protein as a Fine Regulator of the Interaction between HBc and Nucleic Acid.Viruses. 2020 Jul 8;12(7):738. doi: 10.3390/v12070738. Viruses. 2020. PMID: 32650547 Free PMC article. Review.
-
The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: Implications for the development of HBc-targeting antivirals.Antiviral Res. 2018 Jan;149:211-220. doi: 10.1016/j.antiviral.2017.11.015. Epub 2017 Nov 26. Antiviral Res. 2018. PMID: 29183719 Free PMC article. Review.
Cited by
-
Core protein: A pleiotropic keystone in the HBV lifecycle.Antiviral Res. 2015 Sep;121:82-93. doi: 10.1016/j.antiviral.2015.06.020. Epub 2015 Jun 27. Antiviral Res. 2015. PMID: 26129969 Free PMC article. Review.
-
The interface between hepatitis B virus capsid proteins affects self-assembly, pregenomic RNA packaging, and reverse transcription.J Virol. 2015 Mar;89(6):3275-84. doi: 10.1128/JVI.03545-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568211 Free PMC article.
-
A New Role for Capsid Assembly Modulators To Target Mature Hepatitis B Virus Capsids and Prevent Virus Infection.Antimicrob Agents Chemother. 2019 Dec 20;64(1):e01440-19. doi: 10.1128/AAC.01440-19. Print 2019 Dec 20. Antimicrob Agents Chemother. 2019. PMID: 31658963 Free PMC article.
-
A Therapeutic Hepatitis B Virus DNA Vaccine Induces Specific Immune Responses in Mice and Non-Human Primates.Vaccines (Basel). 2021 Aug 29;9(9):969. doi: 10.3390/vaccines9090969. Vaccines (Basel). 2021. PMID: 34579206 Free PMC article.
-
Hepatitis B Virus Core Protein Phosphorylation Sites Affect Capsid Stability and Transient Exposure of the C-terminal Domain.J Biol Chem. 2015 Nov 20;290(47):28584-28593. doi: 10.1074/jbc.M115.678441. Epub 2015 Sep 24. J Biol Chem. 2015. PMID: 26405031 Free PMC article.
References
-
- Shih C. 2012. Chronic hepatitis B and C: basic science to clinical applications. World Scientific, Hackensack, NJ
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

