Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D
- PMID: 24352455
- PMCID: PMC3958080
- DOI: 10.1128/JVI.03228-13
Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D
Erratum in
-
Correction for McGuire et al., diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D.J Virol. 2015 May;89(9):5194. doi: 10.1128/JVI.00341-15. J Virol. 2015. PMID: 25841051 Free PMC article. No abstract available.
Abstract
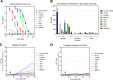
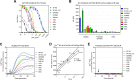
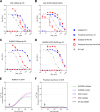
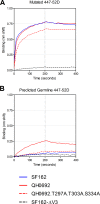
Broadly neutralizing antibodies (bNAbs) against HIV-1 are generated during HIV-1-infection but have not yet been elicited by immunization with recombinant forms of the viral envelope glycoprotein (Env; the target of anti-HIV-1 neutralizing antibodies). A particular type of bNAb targets the CD4-binding site (CD4-BS) region of Env. These antibodies are derived from a limited number of VH/VL genes and can bind to and neutralize diverse HIV-1 strains. Recent reports have demonstrated the limited potential of Env to activate B cells expressing the germline B cell receptor (BCR) forms of anti-CD4-BS bNAbs. A potential reason for the lack of elicitation of anti-CD4-BS bNAbs by Env immunogens is the absence of stimulation of naive B cells expressing the germline BCRs of such antibodies. Several bNAbs have been isolated from HIV-1-infected subjects that target other structurally conserved regions of Env. How frequently Env immunogens stimulate the germline BCRs that give rise to bNAbs that target Env regions other than the CD4-BS is not well understood. Here, we investigated the interactions between diverse Envs and the BCRs of known bNAbs targeting not only the CD4-BS but also conserved elements of the second and third variable Env regions. Our results indicate that Env is generally ineffective in engaging germline BCRs of bNAbs irrespective of their epitope target. Potentially, this is the result of viral evolutionary mechanisms adopted to escape broadly neutralizing antibody responses. Our results also suggest that a single Env capable of activating germline BCRs that target distinct Env epitopes will be very difficult to identify or to design.
Importance: Broadly neutralizing antibodies against HIV-1 are thought to be an important component of the immune responses that a successful vaccine should elicit. Broadly neutralizing antibodies are generated by a subset of those infected by HIV-1, but so far, they have not been generated by immunization with recombinant Envelope (Env, the target of anti-HIV-1 neutralizing antibodies). Here, we provide evidence that the inability of Env to elicit the production of broadly neutralizing antibodies is due to the inability of diverse Envs to engage the germline B cell receptor forms of known broadly neutralizing antibodies.
Figures

Similar articles
-
Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection.Curr Opin HIV AIDS. 2014 May;9(3):210-6. doi: 10.1097/COH.0000000000000057. Curr Opin HIV AIDS. 2014. PMID: 24662931 Free PMC article. Review.
-
Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies.J Exp Med. 2013 Apr 8;210(4):655-63. doi: 10.1084/jem.20122824. Epub 2013 Mar 25. J Exp Med. 2013. PMID: 23530120 Free PMC article.
-
Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs.PLoS Pathog. 2013 Jan;9(1):e1003106. doi: 10.1371/journal.ppat.1003106. Epub 2013 Jan 3. PLoS Pathog. 2013. PMID: 23300456 Free PMC article.
-
HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies.Science. 2014 Dec 12;346(6215):1380-1383. doi: 10.1126/science.1259206. Science. 2014. PMID: 25504724 Free PMC article.
-
Germline-targeting immunogens.Immunol Rev. 2017 Jan;275(1):203-216. doi: 10.1111/imr.12483. Immunol Rev. 2017. PMID: 28133796 Free PMC article. Review.
Cited by
-
Development of a VRC01-class germline targeting immunogen derived from anti-idiotypic antibodies.Cell Rep. 2021 May 4;35(5):109084. doi: 10.1016/j.celrep.2021.109084. Cell Rep. 2021. PMID: 33951425 Free PMC article.
-
Display of the HIV envelope protein at the yeast cell surface for immunogen development.PLoS One. 2018 Oct 18;13(10):e0205756. doi: 10.1371/journal.pone.0205756. eCollection 2018. PLoS One. 2018. PMID: 30335821 Free PMC article.
-
A pathway to HIV-1 neutralization breadth.Nat Med. 2015 Nov;21(11):1246-7. doi: 10.1038/nm.3989. Nat Med. 2015. PMID: 26540383 Free PMC article.
-
Ontogeny-based immunogens for the induction of V2-directed HIV broadly neutralizing antibodies.Immunol Rev. 2017 Jan;275(1):217-229. doi: 10.1111/imr.12501. Immunol Rev. 2017. PMID: 28133797 Free PMC article. Review.
-
Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection.Curr Opin HIV AIDS. 2014 May;9(3):210-6. doi: 10.1097/COH.0000000000000057. Curr Opin HIV AIDS. 2014. PMID: 24662931 Free PMC article. Review.
References
-
- Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769. 10.1128/JVI.02036-08 - DOI - PMC - PubMed
-
- Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636. 10.1128/JVI.01482-09 - DOI - PMC - PubMed
-
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840. 10.1128/JVI.00198-11 - DOI - PMC - PubMed
-
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee D, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. HIV-1 elite neutralizers: individuals with broad and potent neutralizing activity identified using a high throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348. 10.1128/JVI.00110-09 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

