Human enterovirus 71 uncoating captured at atomic resolution
- PMID: 24352461
- PMCID: PMC3957924
- DOI: 10.1128/JVI.03029-13
Human enterovirus 71 uncoating captured at atomic resolution
Abstract
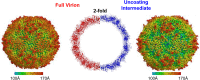
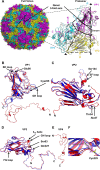
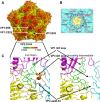
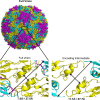
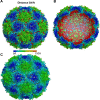
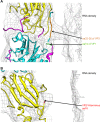
Human enterovirus 71 (EV71) is the major causative agent of severe hand-foot-and-mouth diseases (HFMD) in young children, and structural characterization of EV71 during its life cycle can aid in the development of therapeutics against HFMD. Here, we present the atomic structures of the full virion and an uncoating intermediate of a clinical EV71 C4 strain to illustrate the structural changes in the full virion that lead to the formation of the uncoating intermediate prepared for RNA release. Although the VP1 N-terminal regions observed to penetrate through the junction channel at the quasi-3-fold axis in the uncoating intermediate of coxsackievirus A16 were not observed in the EV71 uncoating intermediate, drastic conformational changes occur in this region, as has been observed in all capsid proteins. Additionally, the RNA genome interacts with the N-terminal extensions of VP1 and residues 32 to 36 of VP3, both of which are situated at the bottom of the junction. These observations highlight the importance of the junction for genome release. Furthermore, EV71 uncoating is associated with apparent rearrangements and expansion around the 2- and 5-fold axes without obvious changes around the 3-fold axes. Therefore, these structures enabled the identification of hot spots for capsid rearrangements, which led to the hypothesis that the protomer interface near the junction and the 2-fold axis permits the opening of large channels for the exit of polypeptides and viral RNA, which is an uncoating mechanism that is likely conserved in enteroviruses.
Importance: Human enterovirus 71 (EV71) is the major causative agent of severe hand-foot-and-mouth diseases (HFMD) in young children. EV71 contains an RNA genome protected by an icosahedral capsid shell. Uncoating is essential in EV71 life cycle, which is characterized by conformational changes in the capsid to facilitate RNA release into host cell. Here we present the atomic structures of the full virion and an uncoating intermediate of a clinical C4 strain of EV71. Structural analysis revealed drastic conformational changes associated with uncoating in all the capsid proteins near the junction at the quasi-3-fold axis and protein-RNA interactions at the bottom of the junction in the uncoating intermediate. Significant capsid rearrangements also occur at the icosahedral 2- and 5-fold axes but not at the 3-fold axis. Taking the results together, we hypothesize that the junction and nearby areas are hot spots for capsid breaches for the exit of polypeptides and viral RNA during uncoating.
Figures



References
-
- Racaniello VR. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

