Combined effects of ankylosing spondylitis-associated ERAP1 polymorphisms outside the catalytic and peptide-binding sites on the processing of natural HLA-B27 ligands
- PMID: 24352655
- PMCID: PMC3924265
- DOI: 10.1074/jbc.M113.529610
Combined effects of ankylosing spondylitis-associated ERAP1 polymorphisms outside the catalytic and peptide-binding sites on the processing of natural HLA-B27 ligands
Abstract
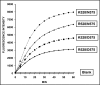
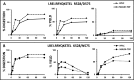
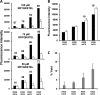
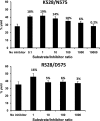
ERAP1 polymorphism involving residues 528 and 575/725 is associated with ankylosing spondylitis among HLA-B27-positive individuals. We used four recombinant variants to address the combined effects of the K528R and D575N polymorphism on the processing of HLA-B27 ligands. The hydrolysis of a fluorogenic substrate, Arg-528/Asp-575 < Lys-528/Asp-575 < Arg-528/Asn-575 < Lys-528/Asn-575, indicated that the relative activity of variants carrying Arg-528 or Lys-528 depends on residue 575. Asp-575 conferred lower activity than Asn-575, but the difference depended on residue 528. The same hierarchy was observed with synthetic precursors of HLA-B27 ligands, but the effects were peptide-dependent. Sometimes the epitope yields were variant-specific at all times. For other peptides, concomitant generation and destruction led to similar epitope amounts with all the variants at long, but not at short, digestion times. The generation/destruction balance of two related HLA-B27 ligands was analyzed in vitro and in live cells. Their relative yields at long digestion times were comparable with those from HLA-B27-positive cells, suggesting that ERAP1 was a major determinant of the abundance of these peptides in vivo. The hydrolysis of fluorogenic and peptide substrates by an HLA-B27 ligand or a shorter peptide, respectively, was increasingly inhibited as a function of ERAP1 activity, indicating that residues 528 and 575 affect substrate inhibition of ERAP1 trimming. The significant and complex effects of co-occurring ERAP1 polymorphisms on multiple HLA-B27 ligands, and their potential to alter the immunological and pathogenetic features of HLA-B27 as a function of the ERAP1 context, explain the epistatic association of both molecules in ankylosing spondylitis.
Keywords: Aminopeptidase; Antigen Processing; Arthritis; Major Histocompatibility Complex (MHC); Pathogenesis; Peptides.
Figures



Similar articles
-
Ranking the Contribution of Ankylosing Spondylitis-associated Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) Polymorphisms to Shaping the HLA-B*27 Peptidome.Mol Cell Proteomics. 2018 Jul;17(7):1308-1323. doi: 10.1074/mcp.RA117.000565. Epub 2018 Apr 9. Mol Cell Proteomics. 2018. PMID: 29632046 Free PMC article.
-
Separate effects of the ankylosing spondylitis associated ERAP1 and ERAP2 aminopeptidases determine the influence of their combined phenotype on the HLA-B*27 peptidome.J Autoimmun. 2017 May;79:28-38. doi: 10.1016/j.jaut.2016.12.008. Epub 2017 Jan 4. J Autoimmun. 2017. PMID: 28063628
-
ERAP1 in ankylosing spondylitis: genetics, biology and pathogenetic role.Curr Opin Rheumatol. 2013 Jul;25(4):419-25. doi: 10.1097/BOR.0b013e328362042f. Curr Opin Rheumatol. 2013. PMID: 23656713 Review.
-
Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens.Autoimmunity. 2013 Dec;46(8):497-508. doi: 10.3109/08916934.2013.819855. Epub 2013 Sep 13. Autoimmunity. 2013. PMID: 24028501 Free PMC article.
-
ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases.Mol Immunol. 2014 Jan;57(1):12-21. doi: 10.1016/j.molimm.2013.06.012. Epub 2013 Jul 31. Mol Immunol. 2014. PMID: 23916068 Review.
Cited by
-
Common allotypes of ER aminopeptidase 1 have substrate-dependent and highly variable enzymatic properties.J Biol Chem. 2021 Jan-Jun;296:100443. doi: 10.1016/j.jbc.2021.100443. Epub 2021 Feb 20. J Biol Chem. 2021. PMID: 33617882 Free PMC article.
-
Ranking the Contribution of Ankylosing Spondylitis-associated Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) Polymorphisms to Shaping the HLA-B*27 Peptidome.Mol Cell Proteomics. 2018 Jul;17(7):1308-1323. doi: 10.1074/mcp.RA117.000565. Epub 2018 Apr 9. Mol Cell Proteomics. 2018. PMID: 29632046 Free PMC article.
-
An allelic variant in the intergenic region between ERAP1 and ERAP2 correlates with an inverse expression of the two genes.Sci Rep. 2018 Jul 10;8(1):10398. doi: 10.1038/s41598-018-28799-8. Sci Rep. 2018. PMID: 29991817 Free PMC article.
-
Know thy immune self and non-self: Proteomics informs on the expanse of self and non-self, and how and where they arise.Proteomics. 2021 Dec;21(23-24):e2000143. doi: 10.1002/pmic.202000143. Epub 2021 Aug 9. Proteomics. 2021. PMID: 34310018 Free PMC article. Review.
-
An Overview on ERAP Roles in Infectious Diseases.Cells. 2020 Mar 14;9(3):720. doi: 10.3390/cells9030720. Cells. 2020. PMID: 32183384 Free PMC article. Review.
References
-
- York I. A., Chang S. C., Saric T., Keys J. A., Favreau J. M., Goldberg A. L., Rock K. L. (2002) The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat. Immunol. 3, 1177–1184 - PubMed
-
- Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., Goldberg A. L. (2002) An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 - PubMed
-
- Serwold T., Gonzalez F., Kim J., Jacob R., Shastri N. (2002) ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–483 - PubMed
-
- Wellcome Trust Case Control Consortium, Australo-Anglo-American Spondylitis Consortium (TASC), Burton P. R., Clayton D. G., Cardon L. R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D. P., McCarthy M. I., Ouwehand W. H., Samani N. J., Todd J. A., Donnelly P., Barrett J. C., Davison D., Easton D., Evans D. M., Leung H. T., Marchini J. L., Morris A. P., Spencer C. C., Tobin M. D., Attwood A. P., Boorman J. P., Cant B., Everson U., Hussey J. M., Jolley J. D., Knight A. S., Koch K., Meech E., Nutland S., Prowse C. V., Stevens H. E., Taylor N. C., Walters G. R., Walker N. M., Watkins N. A., Winzer T., Jones R. W., McArdle W. L., Ring S. M., Strachan D. P., Pembrey M., Breen G., St Clair D., Caesar S., Gordon-Smith K., Jones L., Fraser C., Green E. K., Grozeva D., Hamshere M. L., Holmans P. A., Jones I. R., Kirov G., Moskivina V., Nikolov I., O'Donovan M. C., Owen M. J., Collier D. A., Elkin A., Farmer A., Williamson R., McGuffin P., Young A. H., Ferrier I. N., Ball S. G., Balmforth A. J., Barrett J. H., Bishop T. D., Iles M. M., Maqbool A., Yuldasheva N., Hall A. S., Braund P. S., Dixon R. J., Mangino M., Stevens S., Thompson J. R., Bredin F., Tremelling M., Parkes M., Drummond H., Lees C. W., Nimmo E. R., Satsangi J., Fisher S. A., Forbes A., Lewis C. M., Onnie C. M., Prescott N. J., Sanderson J., Matthew C. G., Barbour J., Mohiuddin M. K., Todhunter C. E., Mansfield J. C., Ahmad T., Cummings F. R., Jewell D. P., Webster J., Brown M. J., Lathrop M. G., Connell J., Dominiczak A., Marcano C. A., Burke B., Dobson R., Gungadoo J., Lee K. L., Munroe P. B., Newhouse S. J., Onipinla A., Wallace C., Xue M., Caulfield M., Farrall M., Barton A., Biologics in RA Genetics and Genomics Study Syndicate (BRAGGS) Steering Committee, Bruce I. N., Donovan H., Eyre S., Gilbert P. D., Hilder S. L., Hinks A. M., John S. L., Potter C., Silman A. J., Symmons D. P., Thomson W., Worthington J., Dunger D. B., Widmer B., Frayling T. M., Freathy R. M., Lango H., Perry J. R., Shields B. M., Weedon M. N., Hattersley A. T., Hitman G. A., Walker M., Elliott K. S., Groves C. J., Lindgren C. M., Rayner N. W., Timpson N. J., Zeggini E., Newport M., Sirugo G., Lyons E., Vannberg F., Hill A. V., Bradbury L. A., Farrar C., Pointon J. J., Wordsworth P., Brown M. A., Franklyn J. A., Heward J. M., Simmonds M. J., Gough S. C., Seal S.; Breast Cancer Susceptibility Collaboration (UK), Stratton M. R., Rahman N., Ban M., Goris A., Sawcer S. J., Compston A., Conway D., Jallow M., Newport M., Sirugo G., Rockett K. A., Bumpstead S. J., Chaney A., Downes K., Ghori M. J., Gwilliam R., Hunt S. E., Inouye M., Keniry A., King E., McGinnis R., Potter S., Ravindrarajah R., Whittaker P., Widden C., Withers D., Cardin N. J., Davison D., Ferreira T., Pereira-Gale J., Hallgrimsdo'ttir I. B., Howie B. N., Su Z., Teo Y. Y., Vukcevic D., Bentley D., Brown M. A., Compston A., Farrall M., Hall A. S., Hattersley A. T., Hill A. V., Parkes M., Pembrey M., Stratton M. R., Mitchell S. L., Newby P. R., Brand O. J., Carr-Smith J., Pearce S. H., McGinnis R., Keniry A., Deloukas P., Reveille J. D., Zhou X., Sims A. M., Dowling A., Taylor J., Doan T., Davis J. C., Savage L., Ward M. M., Learch T. L., Weisman M. H., Brown M. (2007) Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

