Automated versus manual sample inoculations in routine clinical microbiology: a performance evaluation of the fully automated InoqulA instrument
- PMID: 24353001
- PMCID: PMC3957760
- DOI: 10.1128/JCM.02341-13
Automated versus manual sample inoculations in routine clinical microbiology: a performance evaluation of the fully automated InoqulA instrument
Abstract
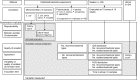
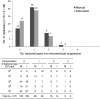
The process of plate streaking has been automated to improve the culture readings, isolation quality, and workflow of microbiology laboratories. However, instruments have not been well evaluated under routine conditions. We aimed to evaluate the performance of the fully automated InoqulA instrument (BD Kiestra B.V., The Netherlands) in the automated seeding of liquid specimens and samples collected using swabs with transport medium. We compared manual and automated methods according to the (i) within-run reproducibility using Escherichia coli-calibrated suspensions, (ii) intersample contamination using a series of alternating sterile broths and broths with >10(5) CFU/ml of either E. coli or Proteus mirabilis, (iii) isolation quality with standardized mixed bacterial suspensions of diverse complexity and a 4-category standardized scale (very poor, poor, fair to good, or excellent), and (iv) agreement of the results obtained from 244 clinical specimens. By involving 15 technicians in the latter part of the comparative study, we estimated the variability in the culture quality at the level of the laboratory team. The instrument produced satisfactory reproducibility with no sample cross-contamination, and it performed better than the manual method, with more colony types recovered and isolated (up to 11% and 17%, respectively). Finally, we showed that the instrument did not shorten the seeding time over short periods of work compared to that for the manual method. Altogether, the instrument improved the quality and standardization of the isolation, thereby contributing to a better overall workflow, shortened the time to results, and provided more accurate results for polymicrobial specimens.
Figures


References
-
- Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. 2012. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J. Clin. Microbiol. 50:2732–2736. 10.1128/JCM.05501-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

