The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types
- PMID: 24353061
- PMCID: PMC3879818
- DOI: 10.1242/dev.099721
The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types
Abstract
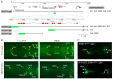
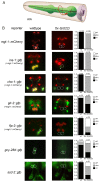
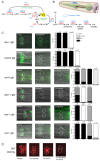
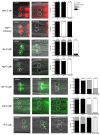
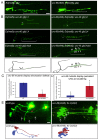
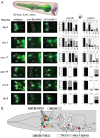
Transcription factors that drive neuron type-specific terminal differentiation programs in the developing nervous system are often expressed in several distinct neuronal cell types, but to what extent they have similar or distinct activities in individual neuronal cell types is generally not well explored. We investigate this problem using, as a starting point, the C. elegans LIM homeodomain transcription factor ttx-3, which acts as a terminal selector to drive the terminal differentiation program of the cholinergic AIY interneuron class. Using a panel of different terminal differentiation markers, including neurotransmitter synthesizing enzymes, neurotransmitter receptors and neuropeptides, we show that ttx-3 also controls the terminal differentiation program of two additional, distinct neuron types, namely the cholinergic AIA interneurons and the serotonergic NSM neurons. We show that the type of differentiation program that is controlled by ttx-3 in different neuron types is specified by a distinct set of collaborating transcription factors. One of the collaborating transcription factors is the POU homeobox gene unc-86, which collaborates with ttx-3 to determine the identity of the serotonergic NSM neurons. unc-86 in turn operates independently of ttx-3 in the anterior ganglion where it collaborates with the ARID-type transcription factor cfi-1 to determine the cholinergic identity of the IL2 sensory and URA motor neurons. In conclusion, transcription factors operate as terminal selectors in distinct combinations in different neuron types, defining neuron type-specific identity features.
Keywords: Caenorhabditis elegans; Homeobox; Neuron differentiation.
Figures


Similar articles
-
Brn3/POU-IV-type POU homeobox genes-Paradigmatic regulators of neuronal identity across phylogeny.Wiley Interdiscip Rev Dev Biol. 2020 Jul;9(4):e374. doi: 10.1002/wdev.374. Epub 2020 Feb 3. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32012462 Review.
-
A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans.Development. 2001 Jun;128(11):1951-69. doi: 10.1242/dev.128.11.1951. Development. 2001. PMID: 11493519
-
The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons.Development. 2002 Aug;129(16):3901-11. doi: 10.1242/dev.129.16.3901. Development. 2002. PMID: 12135927
-
BRN3-type POU Homeobox Genes Maintain the Identity of Mature Postmitotic Neurons in Nematodes and Mice.Curr Biol. 2018 Sep 10;28(17):2813-2823.e2. doi: 10.1016/j.cub.2018.06.045. Epub 2018 Aug 23. Curr Biol. 2018. PMID: 30146154
-
A map of terminal regulators of neuronal identity in Caenorhabditis elegans.Wiley Interdiscip Rev Dev Biol. 2016 Jul;5(4):474-98. doi: 10.1002/wdev.233. Epub 2016 May 2. Wiley Interdiscip Rev Dev Biol. 2016. PMID: 27136279 Free PMC article. Review.
Cited by
-
Caenorhabditis elegans sine oculis/SIX-type homeobox genes act as homeotic switches to define neuronal subtype identities.Proc Natl Acad Sci U S A. 2022 Sep 13;119(37):e2206817119. doi: 10.1073/pnas.2206817119. Epub 2022 Sep 6. Proc Natl Acad Sci U S A. 2022. PMID: 36067313 Free PMC article.
-
Maintenance of neuronal identity in C. elegans and beyond: Lessons from transcription and chromatin factors.Semin Cell Dev Biol. 2024 Feb 15;154(Pt A):35-47. doi: 10.1016/j.semcdb.2023.07.001. Epub 2023 Jul 11. Semin Cell Dev Biol. 2024. PMID: 37438210 Free PMC article. Review.
-
A Receptor Tyrosine Kinase Plays Separate Roles in Sensory Integration and Associative Learning in C. elegans.eNeuro. 2019 Aug 13;6(4):ENEURO.0244-18.2019. doi: 10.1523/ENEURO.0244-18.2019. Print 2019 Jul/Aug. eNeuro. 2019. PMID: 31371455 Free PMC article.
-
Forkhead transcription factor FKH-8 cooperates with RFX in the direct regulation of sensory cilia in Caenorhabditis elegans.Elife. 2023 Jul 14;12:e89702. doi: 10.7554/eLife.89702. Elife. 2023. PMID: 37449480 Free PMC article.
-
Isolation of specific neurons from C. elegans larvae for gene expression profiling.PLoS One. 2014 Nov 5;9(11):e112102. doi: 10.1371/journal.pone.0112102. eCollection 2014. PLoS One. 2014. PMID: 25372608 Free PMC article.
References
-
- Albertson D. G., Thomson J. N. (1976). The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. B 275, 299–325 - PubMed
-
- Altun Z. F., Herndon L. A., Crocker C., Lints R., Hall D. H. (eds) (2002-2013). WormAtlas, http://www.wormatlas.org
-
- Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y., Hobert O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969 - PubMed
-
- Aspöck G., Ruvkun G., Bürglin T. R. (2003). The Caenorhabditis elegans ems class homeobox gene ceh-2 is required for M3 pharynx motoneuron function. Development 130, 3369–3378 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

