Fgf10-positive cells represent a progenitor cell population during lung development and postnatally
- PMID: 24353064
- PMCID: PMC3879811
- DOI: 10.1242/dev.099747
Fgf10-positive cells represent a progenitor cell population during lung development and postnatally
Abstract
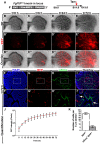
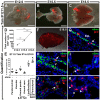
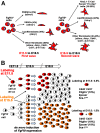
The lung mesenchyme consists of a widely heterogeneous population of cells that play crucial roles during development and homeostasis after birth. These cells belong to myogenic, adipogenic, chondrogenic, neuronal and other lineages. Yet, no clear hierarchy for these lineages has been established. We have previously generated a novel Fgf10(iCre) knock-in mouse line that allows lineage tracing of Fgf10-positive cells during development and postnatally. Using these mice, we hereby demonstrate the presence of two waves of Fgf10 expression during embryonic lung development: the first wave, comprising Fgf10-positive cells residing in the submesothelial mesenchyme at early pseudoglandular stage (as well as their descendants); and the second wave, comprising Fgf10-positive cells from late pseudoglandular stage (as well as their descendants). Our lineage-tracing data reveal that the first wave contributes to the formation of parabronchial and vascular smooth muscle cells as well as lipofibroblasts at later developmental stages, whereas the second wave does not give rise to smooth muscle cells but to lipofibroblasts as well as an Nkx2.1(-) E-Cad(-) Epcam(+) Pro-Spc(+) lineage that requires further in-depth analysis. During alveologenesis, Fgf10-positive cells give rise to lipofibroblasts rather than alveolar myofibroblasts, and during adult life, a subpopulation of Fgf10-expressing cells represents a pool of resident mesenchymal stromal (stem) cells (MSCs) (Cd45(-) Cd31(-) Sca-1(+)). Taken together, we show for the first time that Fgf10-expressing cells represent a pool of mesenchymal progenitors in the embryonic and postnatal lung. Our findings suggest that Fgf10-positive cells could be useful for developing stem cell-based therapies for treating interstitial lung diseases.
Keywords: Fgf10-positive cells; Lineage tracing; Lung mesenchyme; Mouse; Resident MSCs.
Figures





References
-
- Bellusci S., Grindley J., Emoto H., Itoh N., Hogan B. L. (1997). Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124, 4867–4878 - PubMed
-
- Boström H., Willetts K., Pekny M., Levéen P., Lindahl P., Hedstrand H., Pekna M., Hellström M., Gebre-Medhin S., Schalling M., et al. (1996). PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85, 863–873 - PubMed
-
- Crapo J. D., Barry B. E., Gehr P., Bachofen M., Weibel E. R. (1982). Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 126, 332–337 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

