NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins
- PMID: 24353314
- PMCID: PMC3950704
- DOI: 10.1093/nar/gkt1311
NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins
Abstract
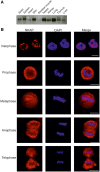
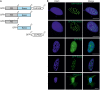
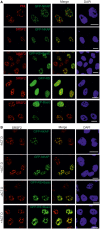
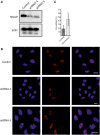
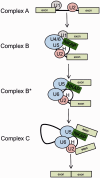
NKAP is a highly conserved protein with roles in transcriptional repression, T-cell development, maturation and acquisition of functional competency and maintenance and survival of adult hematopoietic stem cells. Here we report the novel role of NKAP in splicing. With NKAP-specific antibodies we found that NKAP localizes to nuclear speckles. NKAP has an RS motif at the N-terminus followed by a highly basic domain and a DUF 926 domain at the C-terminal region. Deletion analysis showed that the basic domain is important for speckle localization. In pull-down experiments, we identified RNA-binding proteins, RNA helicases and splicing factors as interaction partners of NKAP, among them FUS/TLS. The FUS/TLS-NKAP interaction takes place through the RS domain of NKAP and the RGG1 and RGG3 domains of FUS/TLS. We analyzed the ability of NKAP to interact with RNA using in vitro splicing assays and found that NKAP bound both spliced messenger RNA (mRNA) and unspliced pre-mRNA. Genome-wide analysis using crosslinking and immunoprecipitation-seq revealed NKAP association with U1, U4 and U5 small nuclear RNA, and we also demonstrated that knockdown of NKAP led to an increase in pre-mRNA percentage. Our results reveal NKAP as nuclear speckle protein with roles in RNA splicing and processing.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

