Models of gene gain and gene loss for probabilistic reconstruction of gene content in the last universal common ancestor of life
- PMID: 24354654
- PMCID: PMC3892064
- DOI: 10.1186/1745-6150-8-32
Models of gene gain and gene loss for probabilistic reconstruction of gene content in the last universal common ancestor of life
Abstract
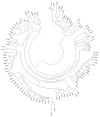
Background: The problem of probabilistic inference of gene content in the last common ancestor of several extant species with completely sequenced genomes is: for each gene that is conserved in all or some of the genomes, assign the probability that its ancestral gene was present in the genome of their last common ancestor.
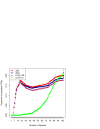
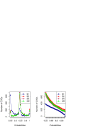
Results: We have developed a family of models of gene gain and gene loss in evolution, and applied the maximum-likelihood approach that uses phylogenetic tree of prokaryotes and the record of orthologous relationships between their genes to infer the gene content of LUCA, the Last Universal Common Ancestor of all currently living cellular organisms. The crucial parameter, the ratio of gene losses and gene gains, was estimated from the data and was higher in models that take account of the number of in-paralogs in genomes than in models that treat gene presences and absences as a binary trait.
Conclusion: While the numbers of genes that are placed confidently into LUCA are similar in the ML methods and in previously published methods that use various parsimony-based approaches, the identities of genes themselves are different. Most of the models of either kind treat the genes found in many existing genomes in a similar way, assigning to them high probabilities of being ancestral ("high ancestrality"). The ML models are more likely than others to assign high ancestrality to the genes that are relatively rare in the present-day genomes.
Figures

References
-
- Mushegian A. Gene content of LUCA, the last universal common ancestor. Front Biosci. 2008;8:4657–66. - PubMed
-
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;8:2. doi: 10.1186/1471-2148-3-2. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

