Cap-binding complex (CBC)
- PMID: 24354960
- PMCID: PMC3901397
- DOI: 10.1042/BJ20131214
Cap-binding complex (CBC)
Erratum in
- Biochem J. 2014 Feb 15;458(1):185
Abstract
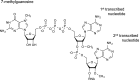
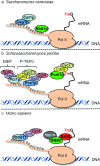
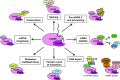
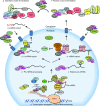
The 7mG (7-methylguanosine cap) formed on mRNA is fundamental to eukaryotic gene expression. Protein complexes recruited to 7mG mediate key processing events throughout the lifetime of the transcript. One of the most important mediators of 7mG functions is CBC (cap-binding complex). CBC has a key role in several gene expression mechanisms, including transcription, splicing, transcript export and translation. Gene expression can be regulated by signalling pathways which influence CBC function. The aim of the present review is to discuss the mechanisms by which CBC mediates and co-ordinates multiple gene expression events.
Figures
References
-
- Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. - PubMed
-
- Topisirovic I., Svitkin Y. V, Sonenberg N., Shatkin A. J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:277–298. - PubMed
-
- Shatkin A. J., Manley J. L. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:838–842. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

